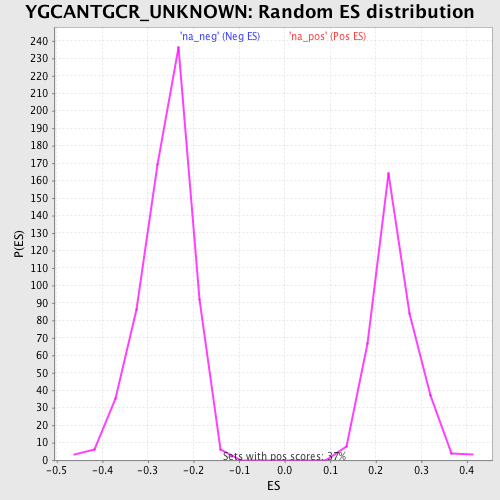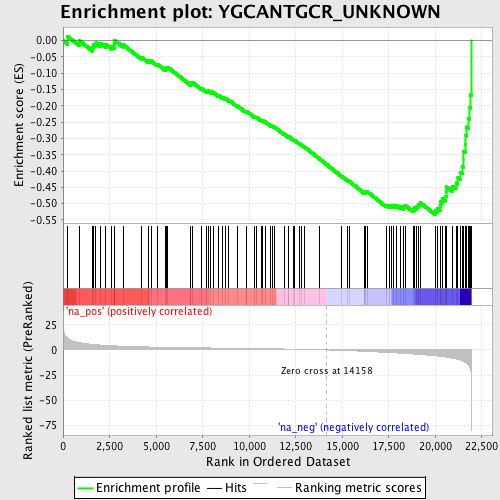
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | YGCANTGCR_UNKNOWN |
| Enrichment Score (ES) | -0.53320014 |
| Normalized Enrichment Score (NES) | -2.0469682 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.003852055 |
| FWER p-Value | 0.0060 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF5A | 229 | 11.555 | 0.0140 | No | ||
| 2 | NOL5A | 873 | 7.320 | 0.0001 | No | ||
| 3 | MIR16 | 1583 | 5.437 | -0.0208 | No | ||
| 4 | SFRS9 | 1650 | 5.318 | -0.0125 | No | ||
| 5 | ASCL2 | 1766 | 5.135 | -0.0069 | No | ||
| 6 | CENTG2 | 2017 | 4.785 | -0.0082 | No | ||
| 7 | EIF4G2 | 2275 | 4.447 | -0.0105 | No | ||
| 8 | HGF | 2609 | 4.099 | -0.0171 | No | ||
| 9 | APP | 2740 | 3.986 | -0.0146 | No | ||
| 10 | SLC39A6 | 2761 | 3.977 | -0.0070 | No | ||
| 11 | NRXN1 | 2777 | 3.968 | 0.0007 | No | ||
| 12 | SLC9A3R2 | 3219 | 3.641 | -0.0118 | No | ||
| 13 | ARHGAP5 | 4207 | 3.039 | -0.0505 | No | ||
| 14 | PHEX | 4578 | 2.883 | -0.0613 | No | ||
| 15 | OLFML3 | 4725 | 2.825 | -0.0620 | No | ||
| 16 | SARS | 5066 | 2.697 | -0.0718 | No | ||
| 17 | KCNS3 | 5499 | 2.556 | -0.0862 | No | ||
| 18 | ASCL1 | 5545 | 2.543 | -0.0828 | No | ||
| 19 | NRXN3 | 5617 | 2.522 | -0.0807 | No | ||
| 20 | FGFR1 | 6855 | 2.152 | -0.1328 | No | ||
| 21 | KPNA1 | 6863 | 2.150 | -0.1285 | No | ||
| 22 | KCNA1 | 6961 | 2.128 | -0.1285 | No | ||
| 23 | PFTK1 | 7459 | 1.997 | -0.1470 | No | ||
| 24 | GAB2 | 7726 | 1.924 | -0.1551 | No | ||
| 25 | MAT2A | 7787 | 1.908 | -0.1538 | No | ||
| 26 | BMPR2 | 7939 | 1.874 | -0.1567 | No | ||
| 27 | SRGAP2 | 8054 | 1.846 | -0.1580 | No | ||
| 28 | PAQR9 | 8348 | 1.780 | -0.1676 | No | ||
| 29 | SYT12 | 8558 | 1.729 | -0.1735 | No | ||
| 30 | LSM11 | 8733 | 1.683 | -0.1779 | No | ||
| 31 | TMEM14A | 8897 | 1.648 | -0.1819 | No | ||
| 32 | IL1RAPL1 | 9371 | 1.536 | -0.2003 | No | ||
| 33 | FREQ | 9841 | 1.425 | -0.2187 | No | ||
| 34 | NLGN3 | 9862 | 1.421 | -0.2166 | No | ||
| 35 | ELOVL4 | 10303 | 1.311 | -0.2340 | No | ||
| 36 | IGSF4D | 10374 | 1.292 | -0.2344 | No | ||
| 37 | MAGED1 | 10645 | 1.227 | -0.2442 | No | ||
| 38 | LRFN5 | 10703 | 1.213 | -0.2442 | No | ||
| 39 | CLSTN1 | 10864 | 1.170 | -0.2490 | No | ||
| 40 | PDCL3 | 11160 | 1.094 | -0.2602 | No | ||
| 41 | SFXN5 | 11255 | 1.068 | -0.2623 | No | ||
| 42 | NNAT | 11369 | 1.033 | -0.2652 | No | ||
| 43 | CALM2 | 11871 | 0.882 | -0.2863 | No | ||
| 44 | PNMA3 | 12111 | 0.808 | -0.2955 | No | ||
| 45 | ACCN1 | 12120 | 0.806 | -0.2942 | No | ||
| 46 | AHI1 | 12368 | 0.732 | -0.3039 | No | ||
| 47 | EPHB1 | 12428 | 0.716 | -0.3051 | No | ||
| 48 | BAI2 | 12714 | 0.624 | -0.3168 | No | ||
| 49 | VSNL1 | 12803 | 0.586 | -0.3196 | No | ||
| 50 | TRIM9 | 12990 | 0.516 | -0.3270 | No | ||
| 51 | ZDHHC15 | 13763 | 0.190 | -0.3619 | No | ||
| 52 | GSK3A | 14949 | -0.501 | -0.4151 | No | ||
| 53 | KCNN2 | 15254 | -0.711 | -0.4275 | No | ||
| 54 | HMGCLL1 | 15367 | -0.781 | -0.4310 | No | ||
| 55 | KLC2 | 16187 | -1.398 | -0.4655 | No | ||
| 56 | STATIP1 | 16201 | -1.408 | -0.4631 | No | ||
| 57 | EWSR1 | 16252 | -1.451 | -0.4623 | No | ||
| 58 | TOPORS | 16344 | -1.520 | -0.4632 | No | ||
| 59 | CALM3 | 17354 | -2.438 | -0.5042 | No | ||
| 60 | PLEKHA1 | 17535 | -2.637 | -0.5069 | No | ||
| 61 | GANAB | 17646 | -2.733 | -0.5061 | No | ||
| 62 | TACC2 | 17778 | -2.868 | -0.5060 | No | ||
| 63 | USP8 | 17898 | -2.998 | -0.5051 | No | ||
| 64 | MNT | 18126 | -3.246 | -0.5086 | No | ||
| 65 | ADNP | 18312 | -3.434 | -0.5098 | No | ||
| 66 | PSAP | 18373 | -3.506 | -0.5051 | No | ||
| 67 | MBTPS2 | 18817 | -4.020 | -0.5169 | No | ||
| 68 | CLTC | 18886 | -4.131 | -0.5112 | No | ||
| 69 | DDX6 | 19013 | -4.300 | -0.5079 | No | ||
| 70 | CAST | 19103 | -4.424 | -0.5025 | No | ||
| 71 | STX16 | 19183 | -4.553 | -0.4965 | No | ||
| 72 | UVRAG | 19986 | -5.842 | -0.5208 | Yes | ||
| 73 | ANXA6 | 20116 | -6.100 | -0.5138 | Yes | ||
| 74 | MAP2K7 | 20261 | -6.417 | -0.5068 | Yes | ||
| 75 | SEMA7A | 20279 | -6.443 | -0.4939 | Yes | ||
| 76 | PCM1 | 20393 | -6.701 | -0.4848 | Yes | ||
| 77 | MGEA5 | 20549 | -7.076 | -0.4769 | Yes | ||
| 78 | FBXO11 | 20597 | -7.205 | -0.4638 | Yes | ||
| 79 | GBF1 | 20604 | -7.224 | -0.4487 | Yes | ||
| 80 | CPNE1 | 20927 | -8.271 | -0.4459 | Yes | ||
| 81 | NONO | 21115 | -8.993 | -0.4354 | Yes | ||
| 82 | CAMK1D | 21215 | -9.463 | -0.4199 | Yes | ||
| 83 | CHD2 | 21351 | -10.205 | -0.4044 | Yes | ||
| 84 | SYNJ1 | 21447 | -10.859 | -0.3857 | Yes | ||
| 85 | GSPT1 | 21530 | -11.532 | -0.3650 | Yes | ||
| 86 | PURA | 21531 | -11.535 | -0.3406 | Yes | ||
| 87 | HS3ST3B1 | 21607 | -12.420 | -0.3177 | Yes | ||
| 88 | MLL5 | 21645 | -12.951 | -0.2919 | Yes | ||
| 89 | CTCF | 21662 | -13.154 | -0.2647 | Yes | ||
| 90 | RTN3 | 21765 | -14.822 | -0.2380 | Yes | ||
| 91 | DGKD | 21846 | -17.377 | -0.2048 | Yes | ||
| 92 | VAMP1 | 21887 | -19.080 | -0.1661 | Yes | ||
| 93 | FGF13 | 21947 | -79.619 | 0.0000 | Yes |