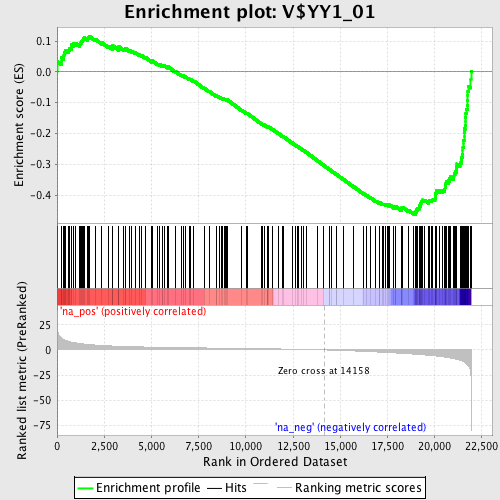
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$YY1_01 |
| Enrichment Score (ES) | -0.4631406 |
| Normalized Enrichment Score (NES) | -1.9236225 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006033808 |
| FWER p-Value | 0.033 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PBX3 | 2 | 31.298 | 0.0329 | No | ||
| 2 | STAG1 | 231 | 11.525 | 0.0346 | No | ||
| 3 | NLN | 235 | 11.513 | 0.0466 | No | ||
| 4 | SLC25A4 | 361 | 10.176 | 0.0516 | No | ||
| 5 | CYR61 | 367 | 10.135 | 0.0621 | No | ||
| 6 | PEX7 | 418 | 9.618 | 0.0700 | No | ||
| 7 | SFRS2 | 597 | 8.452 | 0.0707 | No | ||
| 8 | PCBP4 | 645 | 8.231 | 0.0772 | No | ||
| 9 | RBMX | 770 | 7.690 | 0.0796 | No | ||
| 10 | DSTN | 775 | 7.673 | 0.0876 | No | ||
| 11 | SYNCRIP | 870 | 7.330 | 0.0910 | No | ||
| 12 | SOX14 | 996 | 6.888 | 0.0925 | No | ||
| 13 | TPM1 | 1198 | 6.300 | 0.0899 | No | ||
| 14 | MATR3 | 1259 | 6.136 | 0.0936 | No | ||
| 15 | CXADR | 1271 | 6.114 | 0.0996 | No | ||
| 16 | E2F3 | 1330 | 5.964 | 0.1032 | No | ||
| 17 | INPPL1 | 1375 | 5.849 | 0.1074 | No | ||
| 18 | CD68 | 1426 | 5.762 | 0.1111 | No | ||
| 19 | WDR34 | 1613 | 5.384 | 0.1083 | No | ||
| 20 | PWWP2 | 1645 | 5.325 | 0.1125 | No | ||
| 21 | ADK | 1692 | 5.260 | 0.1159 | No | ||
| 22 | DDX3X | 2012 | 4.789 | 0.1063 | No | ||
| 23 | CDH6 | 2368 | 4.339 | 0.0946 | No | ||
| 24 | IL17B | 2734 | 3.987 | 0.0820 | No | ||
| 25 | SFRS3 | 2925 | 3.854 | 0.0774 | No | ||
| 26 | FAF1 | 2945 | 3.834 | 0.0805 | No | ||
| 27 | ZFHX1B | 2954 | 3.830 | 0.0842 | No | ||
| 28 | NETO1 | 3269 | 3.607 | 0.0736 | No | ||
| 29 | OPRM1 | 3274 | 3.605 | 0.0772 | No | ||
| 30 | SPRY4 | 3276 | 3.604 | 0.0810 | No | ||
| 31 | TPM3 | 3543 | 3.410 | 0.0723 | No | ||
| 32 | ATP5G2 | 3603 | 3.359 | 0.0732 | No | ||
| 33 | FHL3 | 3622 | 3.348 | 0.0759 | No | ||
| 34 | SLC25A13 | 3857 | 3.225 | 0.0685 | No | ||
| 35 | TCF4 | 3960 | 3.166 | 0.0672 | No | ||
| 36 | GTF2A1 | 4142 | 3.074 | 0.0621 | No | ||
| 37 | RHOJ | 4368 | 2.967 | 0.0549 | No | ||
| 38 | PRDM10 | 4463 | 2.926 | 0.0537 | No | ||
| 39 | MYO1E | 4668 | 2.849 | 0.0473 | No | ||
| 40 | PLXNB1 | 5024 | 2.711 | 0.0339 | No | ||
| 41 | FLRT3 | 5039 | 2.707 | 0.0361 | No | ||
| 42 | ODZ1 | 5344 | 2.609 | 0.0249 | No | ||
| 43 | DLX1 | 5416 | 2.586 | 0.0243 | No | ||
| 44 | NCAM2 | 5572 | 2.535 | 0.0199 | No | ||
| 45 | LHX9 | 5610 | 2.523 | 0.0209 | No | ||
| 46 | PPP2R2B | 5684 | 2.500 | 0.0201 | No | ||
| 47 | PRIMA1 | 5858 | 2.444 | 0.0148 | No | ||
| 48 | GABRA1 | 5881 | 2.436 | 0.0163 | No | ||
| 49 | MYL1 | 6294 | 2.305 | -0.0002 | No | ||
| 50 | GRIK2 | 6586 | 2.228 | -0.0112 | No | ||
| 51 | POPDC2 | 6670 | 2.204 | -0.0127 | No | ||
| 52 | CALD1 | 6794 | 2.172 | -0.0160 | No | ||
| 53 | PHOX2B | 7001 | 2.119 | -0.0232 | No | ||
| 54 | ZPBP2 | 7053 | 2.105 | -0.0234 | No | ||
| 55 | GFRA3 | 7241 | 2.054 | -0.0298 | No | ||
| 56 | EML4 | 7797 | 1.907 | -0.0533 | No | ||
| 57 | SRGAP2 | 8054 | 1.846 | -0.0631 | No | ||
| 58 | SNAP25 | 8428 | 1.761 | -0.0784 | No | ||
| 59 | SCN3A | 8462 | 1.750 | -0.0780 | No | ||
| 60 | PDE4D | 8596 | 1.720 | -0.0823 | No | ||
| 61 | BNC2 | 8700 | 1.690 | -0.0853 | No | ||
| 62 | ID1 | 8781 | 1.676 | -0.0872 | No | ||
| 63 | CDH20 | 8860 | 1.657 | -0.0890 | No | ||
| 64 | ECEL1 | 8914 | 1.645 | -0.0897 | No | ||
| 65 | GRIN2B | 8982 | 1.630 | -0.0911 | No | ||
| 66 | PDZRN4 | 9008 | 1.623 | -0.0905 | No | ||
| 67 | CACNA2D3 | 9793 | 1.434 | -0.1250 | No | ||
| 68 | ATP6V0B | 10031 | 1.377 | -0.1344 | No | ||
| 69 | CACNB2 | 10063 | 1.368 | -0.1344 | No | ||
| 70 | ZZZ3 | 10839 | 1.178 | -0.1688 | No | ||
| 71 | THRAP4 | 10887 | 1.165 | -0.1697 | No | ||
| 72 | CDH10 | 10973 | 1.144 | -0.1724 | No | ||
| 73 | MYH11 | 11125 | 1.102 | -0.1782 | No | ||
| 74 | LMO6 | 11131 | 1.101 | -0.1773 | No | ||
| 75 | EN1 | 11168 | 1.093 | -0.1778 | No | ||
| 76 | FOXP2 | 11204 | 1.084 | -0.1782 | No | ||
| 77 | RAB8B | 11397 | 1.024 | -0.1860 | No | ||
| 78 | SGTB | 11725 | 0.927 | -0.2000 | No | ||
| 79 | CASQ2 | 11956 | 0.853 | -0.2097 | No | ||
| 80 | DUSP10 | 11980 | 0.847 | -0.2098 | No | ||
| 81 | WNT3 | 12449 | 0.708 | -0.2306 | No | ||
| 82 | EPS8L1 | 12615 | 0.653 | -0.2375 | No | ||
| 83 | HOXC10 | 12756 | 0.605 | -0.2433 | No | ||
| 84 | VSNL1 | 12803 | 0.586 | -0.2448 | No | ||
| 85 | TAL2 | 12959 | 0.530 | -0.2513 | No | ||
| 86 | RALGDS | 13052 | 0.495 | -0.2550 | No | ||
| 87 | HNRPC | 13226 | 0.423 | -0.2625 | No | ||
| 88 | MYF5 | 13806 | 0.168 | -0.2890 | No | ||
| 89 | DAP3 | 14129 | 0.012 | -0.3037 | No | ||
| 90 | DPAGT1 | 14439 | -0.178 | -0.3177 | No | ||
| 91 | KIAA1128 | 14455 | -0.187 | -0.3182 | No | ||
| 92 | ZADH2 | 14541 | -0.240 | -0.3219 | No | ||
| 93 | CCR3 | 14806 | -0.419 | -0.3336 | No | ||
| 94 | CASQ1 | 14822 | -0.430 | -0.3338 | No | ||
| 95 | SEC23B | 15156 | -0.655 | -0.3484 | No | ||
| 96 | HOXD10 | 15719 | -1.027 | -0.3731 | No | ||
| 97 | PRM1 | 16241 | -1.442 | -0.3956 | No | ||
| 98 | ROD1 | 16398 | -1.572 | -0.4011 | No | ||
| 99 | PRKACA | 16598 | -1.752 | -0.4084 | No | ||
| 100 | NCDN | 16895 | -2.025 | -0.4198 | No | ||
| 101 | RHOA | 17075 | -2.199 | -0.4257 | No | ||
| 102 | NGFRAP1 | 17082 | -2.200 | -0.4237 | No | ||
| 103 | RBMS1 | 17267 | -2.356 | -0.4296 | No | ||
| 104 | PRKAB2 | 17321 | -2.408 | -0.4295 | No | ||
| 105 | ACTG2 | 17402 | -2.487 | -0.4306 | No | ||
| 106 | RPIA | 17505 | -2.605 | -0.4325 | No | ||
| 107 | RGS12 | 17578 | -2.676 | -0.4330 | No | ||
| 108 | DNAJB4 | 17619 | -2.710 | -0.4320 | No | ||
| 109 | RAB1B | 17847 | -2.944 | -0.4393 | No | ||
| 110 | HIC2 | 17933 | -3.035 | -0.4400 | No | ||
| 111 | AP1G1 | 17936 | -3.039 | -0.4369 | No | ||
| 112 | UBE4B | 18247 | -3.380 | -0.4476 | No | ||
| 113 | ANKRD17 | 18261 | -3.392 | -0.4446 | No | ||
| 114 | ACTN1 | 18273 | -3.406 | -0.4415 | No | ||
| 115 | ADNP | 18312 | -3.434 | -0.4396 | No | ||
| 116 | WTAP | 18639 | -3.794 | -0.4506 | No | ||
| 117 | AKAP8L | 18883 | -4.129 | -0.4574 | No | ||
| 118 | SNX13 | 19009 | -4.292 | -0.4586 | Yes | ||
| 119 | DDX6 | 19013 | -4.300 | -0.4542 | Yes | ||
| 120 | DDAH1 | 19055 | -4.353 | -0.4515 | Yes | ||
| 121 | RAB1A | 19062 | -4.370 | -0.4472 | Yes | ||
| 122 | CAST | 19103 | -4.424 | -0.4443 | Yes | ||
| 123 | FOS | 19178 | -4.546 | -0.4429 | Yes | ||
| 124 | PCF11 | 19184 | -4.553 | -0.4384 | Yes | ||
| 125 | PPP2R2A | 19189 | -4.568 | -0.4337 | Yes | ||
| 126 | SRF | 19248 | -4.646 | -0.4315 | Yes | ||
| 127 | ZFP91 | 19257 | -4.657 | -0.4269 | Yes | ||
| 128 | PRKAG1 | 19286 | -4.710 | -0.4233 | Yes | ||
| 129 | NFE2L1 | 19329 | -4.781 | -0.4201 | Yes | ||
| 130 | PLAG1 | 19351 | -4.812 | -0.4160 | Yes | ||
| 131 | TLE3 | 19490 | -5.019 | -0.4171 | Yes | ||
| 132 | CHCHD7 | 19703 | -5.341 | -0.4212 | Yes | ||
| 133 | KIT | 19722 | -5.366 | -0.4163 | Yes | ||
| 134 | TCTA | 19851 | -5.571 | -0.4163 | Yes | ||
| 135 | FBXW7 | 19916 | -5.698 | -0.4133 | Yes | ||
| 136 | CREM | 20025 | -5.922 | -0.4120 | Yes | ||
| 137 | GTF3C2 | 20038 | -5.958 | -0.4062 | Yes | ||
| 138 | KIF1B | 20048 | -5.972 | -0.4004 | Yes | ||
| 139 | UBE3A | 20067 | -6.019 | -0.3948 | Yes | ||
| 140 | NR2C2 | 20097 | -6.071 | -0.3898 | Yes | ||
| 141 | CTDSP1 | 20120 | -6.107 | -0.3843 | Yes | ||
| 142 | CHM | 20282 | -6.448 | -0.3849 | Yes | ||
| 143 | SRRM2 | 20426 | -6.795 | -0.3843 | Yes | ||
| 144 | TAZ | 20529 | -7.027 | -0.3816 | Yes | ||
| 145 | FBXW11 | 20554 | -7.084 | -0.3752 | Yes | ||
| 146 | SSH2 | 20584 | -7.159 | -0.3690 | Yes | ||
| 147 | GBF1 | 20604 | -7.224 | -0.3622 | Yes | ||
| 148 | PHF6 | 20621 | -7.281 | -0.3553 | Yes | ||
| 149 | FUS | 20756 | -7.663 | -0.3534 | Yes | ||
| 150 | FALZ | 20806 | -7.846 | -0.3473 | Yes | ||
| 151 | TRAF3 | 20854 | -7.990 | -0.3411 | Yes | ||
| 152 | RELA | 21022 | -8.671 | -0.3396 | Yes | ||
| 153 | GNAI2 | 21032 | -8.712 | -0.3308 | Yes | ||
| 154 | EPC1 | 21088 | -8.882 | -0.3240 | Yes | ||
| 155 | NRAS | 21157 | -9.167 | -0.3174 | Yes | ||
| 156 | RNF44 | 21178 | -9.248 | -0.3086 | Yes | ||
| 157 | CD4 | 21188 | -9.300 | -0.2992 | Yes | ||
| 158 | CHD2 | 21351 | -10.205 | -0.2958 | Yes | ||
| 159 | ING4 | 21408 | -10.587 | -0.2872 | Yes | ||
| 160 | MAP4K5 | 21442 | -10.831 | -0.2773 | Yes | ||
| 161 | ASH1L | 21495 | -11.283 | -0.2678 | Yes | ||
| 162 | ARMCX3 | 21504 | -11.347 | -0.2562 | Yes | ||
| 163 | LUC7L2 | 21506 | -11.357 | -0.2443 | Yes | ||
| 164 | PHF12 | 21543 | -11.631 | -0.2336 | Yes | ||
| 165 | CD3E | 21545 | -11.647 | -0.2214 | Yes | ||
| 166 | SCML4 | 21569 | -11.922 | -0.2099 | Yes | ||
| 167 | RAB22A | 21572 | -11.959 | -0.1973 | Yes | ||
| 168 | PAFAH1B1 | 21585 | -12.135 | -0.1851 | Yes | ||
| 169 | PPP2R5C | 21618 | -12.520 | -0.1733 | Yes | ||
| 170 | TLK2 | 21642 | -12.867 | -0.1608 | Yes | ||
| 171 | MLL5 | 21645 | -12.951 | -0.1472 | Yes | ||
| 172 | CTCF | 21662 | -13.154 | -0.1341 | Yes | ||
| 173 | PHF8 | 21692 | -13.659 | -0.1210 | Yes | ||
| 174 | PFN2 | 21738 | -14.470 | -0.1078 | Yes | ||
| 175 | IHPK2 | 21750 | -14.578 | -0.0929 | Yes | ||
| 176 | LCP2 | 21764 | -14.815 | -0.0779 | Yes | ||
| 177 | CBX4 | 21769 | -14.930 | -0.0623 | Yes | ||
| 178 | YWHAE | 21814 | -16.134 | -0.0473 | Yes | ||
| 179 | EGR3 | 21930 | -24.862 | -0.0264 | Yes | ||
| 180 | EGR2 | 21933 | -25.685 | 0.0006 | Yes |