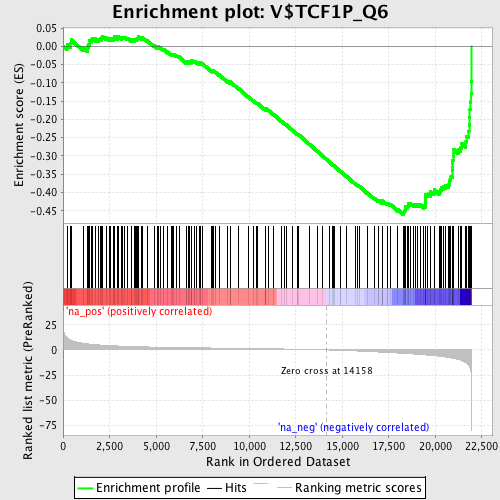
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$TCF1P_Q6 |
| Enrichment Score (ES) | -0.46026188 |
| Normalized Enrichment Score (NES) | -1.9049482 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004299151 |
| FWER p-Value | 0.037 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GMNN | 209 | 11.840 | 0.0047 | No | ||
| 2 | PPAP2A | 401 | 9.812 | 0.0077 | No | ||
| 3 | UCHL5 | 425 | 9.531 | 0.0181 | No | ||
| 4 | TUBA8 | 1077 | 6.637 | -0.0038 | No | ||
| 5 | TFDP2 | 1321 | 5.981 | -0.0077 | No | ||
| 6 | E2F3 | 1330 | 5.964 | -0.0009 | No | ||
| 7 | PTGFRN | 1372 | 5.861 | 0.0043 | No | ||
| 8 | POLA2 | 1425 | 5.762 | 0.0088 | No | ||
| 9 | CD68 | 1426 | 5.762 | 0.0158 | No | ||
| 10 | EEF1A2 | 1510 | 5.580 | 0.0187 | No | ||
| 11 | PCDH9 | 1582 | 5.441 | 0.0220 | No | ||
| 12 | CDKN2C | 1747 | 5.171 | 0.0207 | No | ||
| 13 | STARD7 | 1901 | 4.939 | 0.0196 | No | ||
| 14 | PPARG | 1999 | 4.807 | 0.0209 | No | ||
| 15 | TGIF | 2078 | 4.723 | 0.0230 | No | ||
| 16 | RANGAP1 | 2098 | 4.700 | 0.0278 | No | ||
| 17 | ARX | 2312 | 4.405 | 0.0233 | No | ||
| 18 | PACSIN3 | 2492 | 4.219 | 0.0202 | No | ||
| 19 | ELAVL4 | 2557 | 4.150 | 0.0222 | No | ||
| 20 | PAX6 | 2693 | 4.022 | 0.0209 | No | ||
| 21 | APP | 2740 | 3.986 | 0.0236 | No | ||
| 22 | OAZ3 | 2746 | 3.985 | 0.0281 | No | ||
| 23 | CDKN1A | 2944 | 3.835 | 0.0237 | No | ||
| 24 | HOXC6 | 2951 | 3.830 | 0.0281 | No | ||
| 25 | HS6ST3 | 3148 | 3.695 | 0.0235 | No | ||
| 26 | SEPHS1 | 3199 | 3.656 | 0.0256 | No | ||
| 27 | ADRA2C | 3314 | 3.569 | 0.0247 | No | ||
| 28 | EPHA7 | 3458 | 3.457 | 0.0223 | No | ||
| 29 | PLK1 | 3661 | 3.326 | 0.0170 | No | ||
| 30 | ART5 | 3690 | 3.315 | 0.0197 | No | ||
| 31 | BRUNOL4 | 3841 | 3.234 | 0.0167 | No | ||
| 32 | TYRO3 | 3877 | 3.216 | 0.0190 | No | ||
| 33 | UXT | 3932 | 3.184 | 0.0203 | No | ||
| 34 | PLCD3 | 3975 | 3.161 | 0.0222 | No | ||
| 35 | FZD1 | 4023 | 3.131 | 0.0238 | No | ||
| 36 | PPP2R5D | 4047 | 3.115 | 0.0265 | No | ||
| 37 | ARHGAP5 | 4207 | 3.039 | 0.0229 | No | ||
| 38 | PGAM2 | 4255 | 3.017 | 0.0244 | No | ||
| 39 | RHOD | 4517 | 2.908 | 0.0159 | No | ||
| 40 | ATP2B3 | 4886 | 2.761 | 0.0023 | No | ||
| 41 | PCDH10 | 5086 | 2.690 | -0.0036 | No | ||
| 42 | HOXA4 | 5107 | 2.683 | -0.0013 | No | ||
| 43 | NLGN2 | 5235 | 2.643 | -0.0039 | No | ||
| 44 | NEO1 | 5391 | 2.596 | -0.0079 | No | ||
| 45 | NRXN3 | 5617 | 2.522 | -0.0152 | No | ||
| 46 | TEF | 5819 | 2.458 | -0.0215 | No | ||
| 47 | GATA6 | 5901 | 2.430 | -0.0223 | No | ||
| 48 | EFNA5 | 5947 | 2.415 | -0.0214 | No | ||
| 49 | ARHGAP26 | 6089 | 2.368 | -0.0251 | No | ||
| 50 | PIK3R1 | 6230 | 2.327 | -0.0287 | No | ||
| 51 | BMP10 | 6617 | 2.219 | -0.0438 | No | ||
| 52 | PITX2 | 6622 | 2.217 | -0.0413 | No | ||
| 53 | SEMA3A | 6731 | 2.187 | -0.0436 | No | ||
| 54 | TGFB3 | 6735 | 2.186 | -0.0411 | No | ||
| 55 | NPPC | 6795 | 2.171 | -0.0412 | No | ||
| 56 | GRIA3 | 6891 | 2.143 | -0.0430 | No | ||
| 57 | HOXC4 | 6893 | 2.143 | -0.0404 | No | ||
| 58 | DPM3 | 6895 | 2.143 | -0.0379 | No | ||
| 59 | ZPBP2 | 7053 | 2.105 | -0.0426 | No | ||
| 60 | CENPF | 7069 | 2.101 | -0.0407 | No | ||
| 61 | KITLG | 7182 | 2.070 | -0.0434 | No | ||
| 62 | TBX3 | 7307 | 2.036 | -0.0466 | No | ||
| 63 | COL18A1 | 7363 | 2.021 | -0.0467 | No | ||
| 64 | GRIK1 | 7382 | 2.017 | -0.0451 | No | ||
| 65 | STAG2 | 7493 | 1.988 | -0.0478 | No | ||
| 66 | FOXF2 | 7953 | 1.871 | -0.0666 | No | ||
| 67 | ADCY8 | 8040 | 1.848 | -0.0683 | No | ||
| 68 | SRGAP2 | 8054 | 1.846 | -0.0667 | No | ||
| 69 | EMX2 | 8181 | 1.818 | -0.0703 | No | ||
| 70 | MRGPRF | 8382 | 1.774 | -0.0773 | No | ||
| 71 | DUSP8 | 8849 | 1.661 | -0.0967 | No | ||
| 72 | HOXA11 | 8978 | 1.631 | -0.1007 | No | ||
| 73 | JPH1 | 8980 | 1.630 | -0.0987 | No | ||
| 74 | USP2 | 8988 | 1.629 | -0.0971 | No | ||
| 75 | TRIB1 | 9436 | 1.520 | -0.1158 | No | ||
| 76 | SLC30A3 | 9934 | 1.402 | -0.1369 | No | ||
| 77 | EREG | 10245 | 1.323 | -0.1496 | No | ||
| 78 | NAP1L5 | 10416 | 1.283 | -0.1558 | No | ||
| 79 | FGF9 | 10452 | 1.274 | -0.1559 | No | ||
| 80 | LRRTM4 | 10859 | 1.171 | -0.1731 | No | ||
| 81 | BHLHB3 | 10863 | 1.170 | -0.1719 | No | ||
| 82 | ESRRG | 10869 | 1.169 | -0.1707 | No | ||
| 83 | PAX3 | 10874 | 1.167 | -0.1695 | No | ||
| 84 | ATP6V0A4 | 11016 | 1.130 | -0.1746 | No | ||
| 85 | GPR87 | 11306 | 1.053 | -0.1866 | No | ||
| 86 | CPZ | 11737 | 0.922 | -0.2052 | No | ||
| 87 | FOXA2 | 11905 | 0.869 | -0.2119 | No | ||
| 88 | DUSP10 | 11980 | 0.847 | -0.2142 | No | ||
| 89 | GPX1 | 12331 | 0.745 | -0.2294 | No | ||
| 90 | PRDX2 | 12589 | 0.661 | -0.2404 | No | ||
| 91 | MNAT1 | 12604 | 0.656 | -0.2403 | No | ||
| 92 | IL12B | 12666 | 0.637 | -0.2423 | No | ||
| 93 | TAL1 | 13229 | 0.422 | -0.2676 | No | ||
| 94 | CILP2 | 13690 | 0.229 | -0.2885 | No | ||
| 95 | TNNC1 | 13931 | 0.117 | -0.2993 | No | ||
| 96 | SCGN | 14293 | -0.089 | -0.3158 | No | ||
| 97 | SLC10A2 | 14491 | -0.216 | -0.3246 | No | ||
| 98 | ZADH2 | 14541 | -0.240 | -0.3266 | No | ||
| 99 | GMPPB | 14557 | -0.251 | -0.3269 | No | ||
| 100 | SCNN1A | 14906 | -0.481 | -0.3423 | No | ||
| 101 | CCL20 | 14911 | -0.485 | -0.3419 | No | ||
| 102 | FXYD5 | 15239 | -0.704 | -0.3561 | No | ||
| 103 | BLCAP | 15697 | -1.011 | -0.3759 | No | ||
| 104 | EDN1 | 15814 | -1.087 | -0.3799 | No | ||
| 105 | DEXI | 15911 | -1.162 | -0.3829 | No | ||
| 106 | NPAT | 16352 | -1.531 | -0.4013 | No | ||
| 107 | GGA1 | 16746 | -1.880 | -0.4171 | No | ||
| 108 | EPN1 | 16935 | -2.054 | -0.4232 | No | ||
| 109 | PER1 | 16968 | -2.093 | -0.4222 | No | ||
| 110 | COL15A1 | 17143 | -2.256 | -0.4274 | No | ||
| 111 | CDYL | 17157 | -2.270 | -0.4253 | No | ||
| 112 | MTIF2 | 17169 | -2.282 | -0.4231 | No | ||
| 113 | HNRPL | 17404 | -2.490 | -0.4308 | No | ||
| 114 | HIF1A | 17452 | -2.538 | -0.4299 | No | ||
| 115 | AMMECR1 | 17565 | -2.661 | -0.4318 | No | ||
| 116 | ITPKC | 17989 | -3.094 | -0.4475 | No | ||
| 117 | GLTP | 18267 | -3.397 | -0.4562 | Yes | ||
| 118 | ADNP | 18312 | -3.434 | -0.4541 | Yes | ||
| 119 | PIP5K2B | 18340 | -3.463 | -0.4511 | Yes | ||
| 120 | ACIN1 | 18367 | -3.498 | -0.4481 | Yes | ||
| 121 | HMGCS1 | 18408 | -3.543 | -0.4457 | Yes | ||
| 122 | LMO4 | 18411 | -3.547 | -0.4415 | Yes | ||
| 123 | THRAP1 | 18415 | -3.552 | -0.4374 | Yes | ||
| 124 | HNRPUL1 | 18530 | -3.691 | -0.4381 | Yes | ||
| 125 | NCOA5 | 18547 | -3.706 | -0.4344 | Yes | ||
| 126 | PHF13 | 18563 | -3.718 | -0.4306 | Yes | ||
| 127 | TMIE | 18664 | -3.830 | -0.4306 | Yes | ||
| 128 | ATM | 18851 | -4.077 | -0.4342 | Yes | ||
| 129 | FIP1L1 | 18927 | -4.184 | -0.4326 | Yes | ||
| 130 | SLC9A6 | 19052 | -4.342 | -0.4331 | Yes | ||
| 131 | FOS | 19178 | -4.546 | -0.4334 | Yes | ||
| 132 | CLPX | 19385 | -4.869 | -0.4370 | Yes | ||
| 133 | LTA | 19451 | -4.972 | -0.4340 | Yes | ||
| 134 | DDX5 | 19454 | -4.978 | -0.4281 | Yes | ||
| 135 | CPEB4 | 19462 | -4.985 | -0.4224 | Yes | ||
| 136 | PIK4CB | 19467 | -4.991 | -0.4166 | Yes | ||
| 137 | SP4 | 19487 | -5.016 | -0.4114 | Yes | ||
| 138 | TLE3 | 19490 | -5.019 | -0.4054 | Yes | ||
| 139 | ARF6 | 19599 | -5.188 | -0.4041 | Yes | ||
| 140 | CLK4 | 19723 | -5.369 | -0.4033 | Yes | ||
| 141 | PLEC1 | 19759 | -5.421 | -0.3984 | Yes | ||
| 142 | RBMS2 | 19930 | -5.732 | -0.3993 | Yes | ||
| 143 | KIF3C | 19941 | -5.764 | -0.3928 | Yes | ||
| 144 | TLK1 | 20203 | -6.276 | -0.3973 | Yes | ||
| 145 | SEMA7A | 20279 | -6.443 | -0.3929 | Yes | ||
| 146 | FYCO1 | 20319 | -6.537 | -0.3869 | Yes | ||
| 147 | SPAG9 | 20419 | -6.779 | -0.3832 | Yes | ||
| 148 | SOST | 20557 | -7.092 | -0.3810 | Yes | ||
| 149 | DLG1 | 20702 | -7.506 | -0.3786 | Yes | ||
| 150 | UBR1 | 20738 | -7.590 | -0.3710 | Yes | ||
| 151 | POLR2A | 20787 | -7.778 | -0.3639 | Yes | ||
| 152 | WAC | 20840 | -7.951 | -0.3567 | Yes | ||
| 153 | ETV6 | 20905 | -8.185 | -0.3498 | Yes | ||
| 154 | PIK3CD | 20906 | -8.191 | -0.3399 | Yes | ||
| 155 | CPNE1 | 20927 | -8.271 | -0.3308 | Yes | ||
| 156 | KLK8 | 20933 | -8.296 | -0.3211 | Yes | ||
| 157 | NRF1 | 20939 | -8.341 | -0.3113 | Yes | ||
| 158 | MTSS1 | 20975 | -8.479 | -0.3027 | Yes | ||
| 159 | TPP2 | 20979 | -8.495 | -0.2926 | Yes | ||
| 160 | BCAS3 | 20982 | -8.502 | -0.2824 | Yes | ||
| 161 | CDC42EP3 | 21234 | -9.551 | -0.2824 | Yes | ||
| 162 | NAB2 | 21372 | -10.315 | -0.2763 | Yes | ||
| 163 | MEF2A | 21429 | -10.782 | -0.2659 | Yes | ||
| 164 | MLL5 | 21645 | -12.951 | -0.2602 | Yes | ||
| 165 | PRKD2 | 21683 | -13.494 | -0.2456 | Yes | ||
| 166 | KLF3 | 21779 | -15.184 | -0.2317 | Yes | ||
| 167 | SP1 | 21811 | -15.984 | -0.2139 | Yes | ||
| 168 | ZBTB4 | 21848 | -17.473 | -0.1945 | Yes | ||
| 169 | MARK2 | 21858 | -17.748 | -0.1735 | Yes | ||
| 170 | JARID2 | 21867 | -18.002 | -0.1522 | Yes | ||
| 171 | BCL11B | 21916 | -21.511 | -0.1285 | Yes | ||
| 172 | SATB1 | 21934 | -28.181 | -0.0953 | Yes | ||
| 173 | FGF13 | 21947 | -79.619 | -0.0000 | Yes |