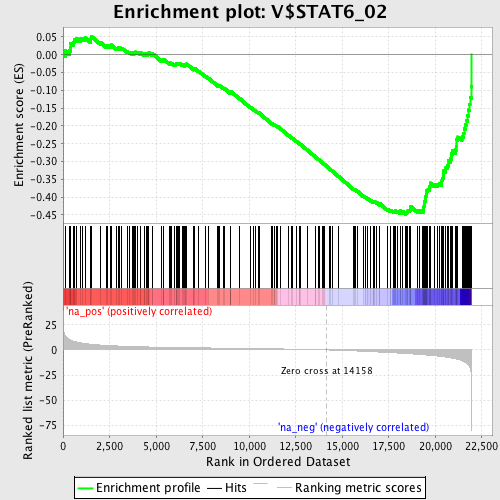
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$STAT6_02 |
| Enrichment Score (ES) | -0.44877896 |
| Normalized Enrichment Score (NES) | -1.8904978 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004112792 |
| FWER p-Value | 0.045 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SNX8 | 112 | 14.216 | 0.0110 | No | ||
| 2 | POLR1D | 356 | 10.207 | 0.0115 | No | ||
| 3 | PDGFRB | 399 | 9.824 | 0.0207 | No | ||
| 4 | ACBD6 | 406 | 9.712 | 0.0315 | No | ||
| 5 | RNF138 | 536 | 8.797 | 0.0356 | No | ||
| 6 | EGFL7 | 590 | 8.483 | 0.0428 | No | ||
| 7 | CRK | 726 | 7.882 | 0.0455 | No | ||
| 8 | ADSL | 928 | 7.118 | 0.0444 | No | ||
| 9 | HMGA1 | 1049 | 6.736 | 0.0466 | No | ||
| 10 | IL7R | 1187 | 6.326 | 0.0475 | No | ||
| 11 | HGFAC | 1465 | 5.652 | 0.0412 | No | ||
| 12 | DHH | 1495 | 5.602 | 0.0462 | No | ||
| 13 | PSCD4 | 1500 | 5.598 | 0.0524 | No | ||
| 14 | SYTL1 | 1992 | 4.815 | 0.0353 | No | ||
| 15 | ATP5E | 2306 | 4.413 | 0.0259 | No | ||
| 16 | SLC31A1 | 2401 | 4.308 | 0.0265 | No | ||
| 17 | FLNC | 2528 | 4.182 | 0.0255 | No | ||
| 18 | NOSTRIN | 2584 | 4.122 | 0.0276 | No | ||
| 19 | NOTCH4 | 2875 | 3.889 | 0.0187 | No | ||
| 20 | ZFHX1B | 2954 | 3.830 | 0.0195 | No | ||
| 21 | LUC7L | 3048 | 3.765 | 0.0195 | No | ||
| 22 | AKT1S1 | 3163 | 3.688 | 0.0185 | No | ||
| 23 | EPHA7 | 3458 | 3.457 | 0.0089 | No | ||
| 24 | SLIT3 | 3578 | 3.377 | 0.0073 | No | ||
| 25 | STAB1 | 3721 | 3.297 | 0.0045 | No | ||
| 26 | A2BP1 | 3775 | 3.268 | 0.0058 | No | ||
| 27 | CENTA1 | 3856 | 3.226 | 0.0058 | No | ||
| 28 | TRPS1 | 3891 | 3.205 | 0.0078 | No | ||
| 29 | TOM1L1 | 4014 | 3.137 | 0.0058 | No | ||
| 30 | PHF15 | 4144 | 3.073 | 0.0034 | No | ||
| 31 | FLT1 | 4151 | 3.068 | 0.0066 | No | ||
| 32 | RHOJ | 4368 | 2.967 | 0.0000 | No | ||
| 33 | MCTS1 | 4390 | 2.959 | 0.0024 | No | ||
| 34 | TWIST1 | 4465 | 2.926 | 0.0024 | No | ||
| 35 | FEV | 4539 | 2.897 | 0.0023 | No | ||
| 36 | HHEX | 4574 | 2.884 | 0.0040 | No | ||
| 37 | POU3F3 | 4612 | 2.868 | 0.0056 | No | ||
| 38 | SPDEF | 4782 | 2.799 | 0.0010 | No | ||
| 39 | ERF | 4798 | 2.793 | 0.0035 | No | ||
| 40 | TCF15 | 5260 | 2.636 | -0.0147 | No | ||
| 41 | CITED4 | 5370 | 2.603 | -0.0168 | No | ||
| 42 | LTC4S | 5394 | 2.594 | -0.0149 | No | ||
| 43 | MEF2C | 5401 | 2.593 | -0.0122 | No | ||
| 44 | DOCK4 | 5715 | 2.491 | -0.0237 | No | ||
| 45 | HTR3B | 5761 | 2.474 | -0.0230 | No | ||
| 46 | DLC1 | 5832 | 2.454 | -0.0234 | No | ||
| 47 | CA3 | 6004 | 2.394 | -0.0286 | No | ||
| 48 | CITED2 | 6065 | 2.376 | -0.0286 | No | ||
| 49 | RUNX3 | 6067 | 2.376 | -0.0260 | No | ||
| 50 | RIN2 | 6085 | 2.370 | -0.0240 | No | ||
| 51 | HOXB4 | 6138 | 2.354 | -0.0237 | No | ||
| 52 | ASPN | 6220 | 2.330 | -0.0248 | No | ||
| 53 | ADCYAP1 | 6258 | 2.317 | -0.0239 | No | ||
| 54 | GREM1 | 6401 | 2.278 | -0.0278 | No | ||
| 55 | MIST | 6472 | 2.260 | -0.0285 | No | ||
| 56 | MTUS1 | 6548 | 2.239 | -0.0294 | No | ||
| 57 | ITGA8 | 6580 | 2.230 | -0.0283 | No | ||
| 58 | HOXB9 | 6593 | 2.227 | -0.0263 | No | ||
| 59 | WNT5A | 6638 | 2.213 | -0.0258 | No | ||
| 60 | ERG | 7011 | 2.116 | -0.0405 | No | ||
| 61 | RAB39 | 7013 | 2.116 | -0.0381 | No | ||
| 62 | VAMP3 | 7083 | 2.094 | -0.0389 | No | ||
| 63 | ARV1 | 7281 | 2.043 | -0.0456 | No | ||
| 64 | SERPINI1 | 7649 | 1.947 | -0.0603 | No | ||
| 65 | CASKIN2 | 7816 | 1.902 | -0.0657 | No | ||
| 66 | VAV3 | 8314 | 1.784 | -0.0866 | No | ||
| 67 | SPI1 | 8324 | 1.783 | -0.0849 | No | ||
| 68 | RNF128 | 8389 | 1.773 | -0.0859 | No | ||
| 69 | PDE4D | 8596 | 1.720 | -0.0934 | No | ||
| 70 | ANGPTL1 | 8668 | 1.699 | -0.0947 | No | ||
| 71 | CRYGB | 8974 | 1.632 | -0.1069 | No | ||
| 72 | GRIN2B | 8982 | 1.630 | -0.1053 | No | ||
| 73 | USP2 | 8988 | 1.629 | -0.1037 | No | ||
| 74 | PDZRN4 | 9008 | 1.623 | -0.1027 | No | ||
| 75 | PAX9 | 9482 | 1.509 | -0.1228 | No | ||
| 76 | SCGB3A1 | 10047 | 1.372 | -0.1471 | No | ||
| 77 | COX17 | 10228 | 1.326 | -0.1539 | No | ||
| 78 | BMP2 | 10357 | 1.296 | -0.1583 | No | ||
| 79 | BDNF | 10515 | 1.257 | -0.1641 | No | ||
| 80 | CLOCK | 10529 | 1.253 | -0.1633 | No | ||
| 81 | CSF3R | 11217 | 1.081 | -0.1936 | No | ||
| 82 | CPA3 | 11268 | 1.064 | -0.1947 | No | ||
| 83 | HOXA10 | 11366 | 1.035 | -0.1980 | No | ||
| 84 | LRRC19 | 11376 | 1.031 | -0.1972 | No | ||
| 85 | LALBA | 11489 | 0.996 | -0.2012 | No | ||
| 86 | TLL1 | 11533 | 0.984 | -0.2021 | No | ||
| 87 | STRC | 11668 | 0.946 | -0.2072 | No | ||
| 88 | LRP2 | 12103 | 0.810 | -0.2262 | No | ||
| 89 | EGF | 12295 | 0.756 | -0.2341 | No | ||
| 90 | GPX1 | 12331 | 0.745 | -0.2349 | No | ||
| 91 | GSC | 12535 | 0.679 | -0.2434 | No | ||
| 92 | SESTD1 | 12712 | 0.625 | -0.2508 | No | ||
| 93 | PSTPIP2 | 12721 | 0.622 | -0.2505 | No | ||
| 94 | CYB561D2 | 12753 | 0.607 | -0.2512 | No | ||
| 95 | PKN3 | 13131 | 0.464 | -0.2680 | No | ||
| 96 | SLC35C1 | 13552 | 0.295 | -0.2870 | No | ||
| 97 | CSF2 | 13564 | 0.288 | -0.2872 | No | ||
| 98 | DCTN2 | 13714 | 0.220 | -0.2938 | No | ||
| 99 | CACNG3 | 13752 | 0.197 | -0.2952 | No | ||
| 100 | NKIRAS2 | 13932 | 0.116 | -0.3033 | No | ||
| 101 | NMNAT2 | 14003 | 0.080 | -0.3065 | No | ||
| 102 | TFEC | 14037 | 0.062 | -0.3079 | No | ||
| 103 | GNGT2 | 14328 | -0.112 | -0.3211 | No | ||
| 104 | GPS2 | 14368 | -0.134 | -0.3227 | No | ||
| 105 | ITPR3 | 14478 | -0.207 | -0.3275 | No | ||
| 106 | ARID1A | 14795 | -0.414 | -0.3416 | No | ||
| 107 | GZMK | 15609 | -0.951 | -0.3779 | No | ||
| 108 | LAT | 15646 | -0.971 | -0.3784 | No | ||
| 109 | HOXA2 | 15700 | -1.015 | -0.3797 | No | ||
| 110 | EDN1 | 15814 | -1.087 | -0.3837 | No | ||
| 111 | EPHA2 | 16134 | -1.353 | -0.3968 | No | ||
| 112 | CD2AP | 16239 | -1.438 | -0.3999 | No | ||
| 113 | LY75 | 16366 | -1.548 | -0.4040 | No | ||
| 114 | CMYA5 | 16542 | -1.697 | -0.4101 | No | ||
| 115 | TNFSF13 | 16654 | -1.800 | -0.4131 | No | ||
| 116 | CTNNB1 | 16702 | -1.841 | -0.4132 | No | ||
| 117 | GNB2 | 16743 | -1.876 | -0.4129 | No | ||
| 118 | LAG3 | 16829 | -1.954 | -0.4146 | No | ||
| 119 | MRPL24 | 17003 | -2.126 | -0.4201 | No | ||
| 120 | GPHN | 17026 | -2.154 | -0.4187 | No | ||
| 121 | SGK | 17430 | -2.515 | -0.4343 | No | ||
| 122 | BZRAP1 | 17576 | -2.674 | -0.4379 | No | ||
| 123 | TANK | 17608 | -2.694 | -0.4363 | No | ||
| 124 | PAX8 | 17776 | -2.867 | -0.4407 | No | ||
| 125 | SLC1A5 | 17823 | -2.923 | -0.4395 | No | ||
| 126 | RAB1B | 17847 | -2.944 | -0.4372 | No | ||
| 127 | FEM1C | 17968 | -3.072 | -0.4392 | No | ||
| 128 | RGS3 | 18102 | -3.221 | -0.4417 | No | ||
| 129 | ZNRF2 | 18103 | -3.222 | -0.4380 | No | ||
| 130 | ANKRD28 | 18262 | -3.392 | -0.4414 | No | ||
| 131 | PDLIM2 | 18423 | -3.564 | -0.4447 | Yes | ||
| 132 | LNX2 | 18457 | -3.608 | -0.4421 | Yes | ||
| 133 | SAP130 | 18507 | -3.667 | -0.4402 | Yes | ||
| 134 | HNRPUL1 | 18530 | -3.691 | -0.4370 | Yes | ||
| 135 | PLCB2 | 18630 | -3.789 | -0.4373 | Yes | ||
| 136 | SDCBP | 18656 | -3.818 | -0.4341 | Yes | ||
| 137 | PDCD10 | 18669 | -3.838 | -0.4303 | Yes | ||
| 138 | RBBP5 | 18688 | -3.864 | -0.4267 | Yes | ||
| 139 | ZF | 19056 | -4.354 | -0.4386 | Yes | ||
| 140 | CNN2 | 19159 | -4.516 | -0.4382 | Yes | ||
| 141 | MBNL1 | 19285 | -4.710 | -0.4385 | Yes | ||
| 142 | ZRANB1 | 19362 | -4.828 | -0.4365 | Yes | ||
| 143 | CENTB2 | 19378 | -4.857 | -0.4317 | Yes | ||
| 144 | KCNQ1 | 19387 | -4.876 | -0.4265 | Yes | ||
| 145 | SLC43A1 | 19399 | -4.890 | -0.4215 | Yes | ||
| 146 | DNAJC7 | 19402 | -4.899 | -0.4160 | Yes | ||
| 147 | JARID1C | 19414 | -4.917 | -0.4109 | Yes | ||
| 148 | FOXO3A | 19458 | -4.980 | -0.4072 | Yes | ||
| 149 | CPEB4 | 19462 | -4.985 | -0.4017 | Yes | ||
| 150 | TRERF1 | 19485 | -5.011 | -0.3970 | Yes | ||
| 151 | TBC1D17 | 19504 | -5.036 | -0.3921 | Yes | ||
| 152 | ZDHHC8 | 19541 | -5.110 | -0.3879 | Yes | ||
| 153 | NFAT5 | 19545 | -5.113 | -0.3822 | Yes | ||
| 154 | CBLB | 19603 | -5.196 | -0.3790 | Yes | ||
| 155 | STAT5A | 19672 | -5.285 | -0.3761 | Yes | ||
| 156 | DOCK11 | 19697 | -5.335 | -0.3711 | Yes | ||
| 157 | PGM2L1 | 19715 | -5.356 | -0.3658 | Yes | ||
| 158 | MPO | 19728 | -5.375 | -0.3602 | Yes | ||
| 159 | HSD11B1 | 19972 | -5.822 | -0.3648 | Yes | ||
| 160 | TUSC4 | 20114 | -6.096 | -0.3643 | Yes | ||
| 161 | ZNF289 | 20197 | -6.262 | -0.3610 | Yes | ||
| 162 | HIVEP1 | 20328 | -6.553 | -0.3595 | Yes | ||
| 163 | TRIM25 | 20355 | -6.612 | -0.3531 | Yes | ||
| 164 | PUM1 | 20383 | -6.680 | -0.3468 | Yes | ||
| 165 | MAP3K3 | 20422 | -6.791 | -0.3408 | Yes | ||
| 166 | SIPA1 | 20428 | -6.800 | -0.3333 | Yes | ||
| 167 | PIM2 | 20446 | -6.848 | -0.3263 | Yes | ||
| 168 | FBXW11 | 20554 | -7.084 | -0.3232 | Yes | ||
| 169 | SEMA4B | 20570 | -7.127 | -0.3157 | Yes | ||
| 170 | CHD6 | 20666 | -7.398 | -0.3117 | Yes | ||
| 171 | RANBP9 | 20690 | -7.468 | -0.3042 | Yes | ||
| 172 | VPS39 | 20693 | -7.474 | -0.2958 | Yes | ||
| 173 | STK4 | 20820 | -7.881 | -0.2927 | Yes | ||
| 174 | TRAF3 | 20854 | -7.990 | -0.2851 | Yes | ||
| 175 | HIF3A | 20869 | -8.066 | -0.2765 | Yes | ||
| 176 | PITPNC1 | 20931 | -8.288 | -0.2699 | Yes | ||
| 177 | FLI1 | 21086 | -8.875 | -0.2669 | Yes | ||
| 178 | TTC13 | 21116 | -8.997 | -0.2580 | Yes | ||
| 179 | SRRM1 | 21127 | -9.050 | -0.2482 | Yes | ||
| 180 | DNAJA2 | 21128 | -9.059 | -0.2379 | Yes | ||
| 181 | CAMK1D | 21215 | -9.463 | -0.2310 | Yes | ||
| 182 | SLC38A2 | 21435 | -10.813 | -0.2288 | Yes | ||
| 183 | ABI3 | 21533 | -11.552 | -0.2201 | Yes | ||
| 184 | PHF12 | 21543 | -11.631 | -0.2073 | Yes | ||
| 185 | PPP2R5C | 21618 | -12.520 | -0.1965 | Yes | ||
| 186 | CTCF | 21662 | -13.154 | -0.1835 | Yes | ||
| 187 | VASP | 21737 | -14.401 | -0.1705 | Yes | ||
| 188 | LCK | 21778 | -15.166 | -0.1551 | Yes | ||
| 189 | MAP4K2 | 21842 | -17.169 | -0.1384 | Yes | ||
| 190 | PRKCB1 | 21866 | -17.985 | -0.1190 | Yes | ||
| 191 | SATB1 | 21934 | -28.181 | -0.0900 | Yes | ||
| 192 | FGF13 | 21947 | -79.619 | -0.0000 | Yes |