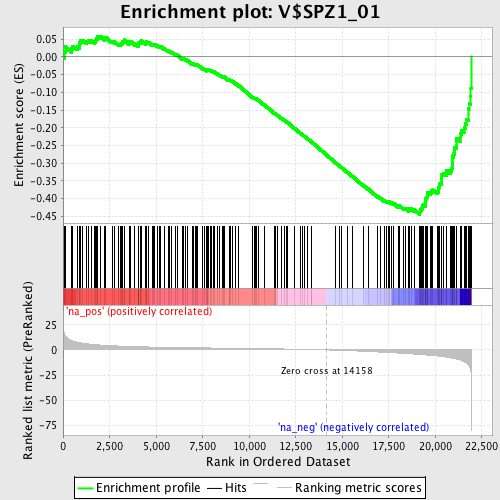
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$SPZ1_01 |
| Enrichment Score (ES) | -0.44448683 |
| Normalized Enrichment Score (NES) | -1.8631972 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004870258 |
| FWER p-Value | 0.072 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MRPS18B | 71 | 15.705 | 0.0144 | No | ||
| 2 | RANBP1 | 110 | 14.273 | 0.0287 | No | ||
| 3 | SNRPD1 | 448 | 9.347 | 0.0238 | No | ||
| 4 | MRPL40 | 524 | 8.847 | 0.0303 | No | ||
| 5 | PABPC1 | 758 | 7.726 | 0.0283 | No | ||
| 6 | CXXC4 | 863 | 7.365 | 0.0318 | No | ||
| 7 | SYNCRIP | 870 | 7.330 | 0.0398 | No | ||
| 8 | CGGBP1 | 922 | 7.135 | 0.0455 | No | ||
| 9 | HMGA1 | 1049 | 6.736 | 0.0473 | No | ||
| 10 | TCF2 | 1272 | 6.112 | 0.0440 | No | ||
| 11 | XPO1 | 1342 | 5.926 | 0.0475 | No | ||
| 12 | PCSK2 | 1505 | 5.586 | 0.0463 | No | ||
| 13 | SMARCC1 | 1696 | 5.252 | 0.0435 | No | ||
| 14 | EFEMP2 | 1749 | 5.169 | 0.0469 | No | ||
| 15 | KPNB1 | 1789 | 5.095 | 0.0509 | No | ||
| 16 | RNF12 | 1828 | 5.042 | 0.0548 | No | ||
| 17 | ILF3 | 1868 | 4.988 | 0.0586 | No | ||
| 18 | YTHDF2 | 1993 | 4.812 | 0.0583 | No | ||
| 19 | SCRT2 | 2240 | 4.484 | 0.0521 | No | ||
| 20 | EIF4G2 | 2275 | 4.447 | 0.0555 | No | ||
| 21 | BCL11A | 2627 | 4.079 | 0.0440 | No | ||
| 22 | IL17B | 2734 | 3.987 | 0.0436 | No | ||
| 23 | SOX3 | 3001 | 3.797 | 0.0357 | No | ||
| 24 | HMGN2 | 3078 | 3.749 | 0.0364 | No | ||
| 25 | NTRK3 | 3133 | 3.707 | 0.0381 | No | ||
| 26 | VCAM1 | 3179 | 3.677 | 0.0402 | No | ||
| 27 | SHANK1 | 3203 | 3.652 | 0.0432 | No | ||
| 28 | SPRY4 | 3276 | 3.604 | 0.0440 | No | ||
| 29 | MRC2 | 3280 | 3.598 | 0.0479 | No | ||
| 30 | TPM3 | 3543 | 3.410 | 0.0397 | No | ||
| 31 | ATF3 | 3548 | 3.407 | 0.0433 | No | ||
| 32 | FHL3 | 3622 | 3.348 | 0.0438 | No | ||
| 33 | HOXB6 | 3860 | 3.223 | 0.0365 | No | ||
| 34 | RBMS3 | 4029 | 3.126 | 0.0323 | No | ||
| 35 | SIX4 | 4050 | 3.114 | 0.0349 | No | ||
| 36 | NR2F2 | 4054 | 3.112 | 0.0382 | No | ||
| 37 | HOXD9 | 4075 | 3.102 | 0.0408 | No | ||
| 38 | GTF2A1 | 4142 | 3.074 | 0.0412 | No | ||
| 39 | HSD3B7 | 4167 | 3.063 | 0.0436 | No | ||
| 40 | STAC2 | 4184 | 3.051 | 0.0463 | No | ||
| 41 | EIF4G1 | 4441 | 2.936 | 0.0378 | No | ||
| 42 | HTF9C | 4461 | 2.927 | 0.0403 | No | ||
| 43 | MYLK | 4466 | 2.925 | 0.0434 | No | ||
| 44 | POU3F3 | 4612 | 2.868 | 0.0399 | No | ||
| 45 | PPP1R1B | 4808 | 2.790 | 0.0341 | No | ||
| 46 | PRRX1 | 4875 | 2.765 | 0.0342 | No | ||
| 47 | PGR | 4901 | 2.755 | 0.0362 | No | ||
| 48 | DQX1 | 5044 | 2.706 | 0.0327 | No | ||
| 49 | POU2F1 | 5182 | 2.660 | 0.0294 | No | ||
| 50 | APBA2BP | 5253 | 2.638 | 0.0291 | No | ||
| 51 | PSCD3 | 5451 | 2.574 | 0.0230 | No | ||
| 52 | BCL7A | 5646 | 2.511 | 0.0169 | No | ||
| 53 | CUL2 | 5718 | 2.490 | 0.0164 | No | ||
| 54 | SLC17A7 | 5840 | 2.450 | 0.0136 | No | ||
| 55 | HYAL2 | 6036 | 2.386 | 0.0074 | No | ||
| 56 | HOXB4 | 6138 | 2.354 | 0.0054 | No | ||
| 57 | GREM1 | 6401 | 2.278 | -0.0041 | No | ||
| 58 | SUMO2 | 6460 | 2.262 | -0.0042 | No | ||
| 59 | HOXB9 | 6593 | 2.227 | -0.0078 | No | ||
| 60 | SOX15 | 6689 | 2.199 | -0.0097 | No | ||
| 61 | IRX5 | 6943 | 2.132 | -0.0189 | No | ||
| 62 | FST | 6992 | 2.121 | -0.0187 | No | ||
| 63 | ADAMTSL1 | 7115 | 2.085 | -0.0220 | No | ||
| 64 | GPM6B | 7193 | 2.065 | -0.0232 | No | ||
| 65 | KCNK2 | 7197 | 2.065 | -0.0210 | No | ||
| 66 | STAG2 | 7493 | 1.988 | -0.0323 | No | ||
| 67 | GLRA1 | 7582 | 1.968 | -0.0341 | No | ||
| 68 | MAGED2 | 7708 | 1.928 | -0.0377 | No | ||
| 69 | WNT10A | 7750 | 1.917 | -0.0374 | No | ||
| 70 | CNTFR | 7783 | 1.910 | -0.0368 | No | ||
| 71 | CASKIN2 | 7816 | 1.902 | -0.0361 | No | ||
| 72 | HOXA9 | 7895 | 1.884 | -0.0375 | No | ||
| 73 | SLITRK1 | 7965 | 1.868 | -0.0386 | No | ||
| 74 | HR | 8080 | 1.842 | -0.0418 | No | ||
| 75 | CDK6 | 8138 | 1.830 | -0.0423 | No | ||
| 76 | CDK5R2 | 8300 | 1.790 | -0.0477 | No | ||
| 77 | SOX10 | 8422 | 1.762 | -0.0513 | No | ||
| 78 | FXYD4 | 8584 | 1.723 | -0.0567 | No | ||
| 79 | FGF17 | 8615 | 1.715 | -0.0562 | No | ||
| 80 | UQCRH | 8656 | 1.702 | -0.0561 | No | ||
| 81 | ECEL1 | 8914 | 1.645 | -0.0661 | No | ||
| 82 | HIRA | 8916 | 1.644 | -0.0643 | No | ||
| 83 | RIMS1 | 8977 | 1.631 | -0.0652 | No | ||
| 84 | PSD | 9091 | 1.602 | -0.0686 | No | ||
| 85 | NPPA | 9272 | 1.559 | -0.0751 | No | ||
| 86 | HDAC8 | 9439 | 1.520 | -0.0810 | No | ||
| 87 | S100A4 | 10186 | 1.337 | -0.1138 | No | ||
| 88 | SHH | 10283 | 1.315 | -0.1167 | No | ||
| 89 | EPN3 | 10315 | 1.307 | -0.1167 | No | ||
| 90 | IGSF4D | 10374 | 1.292 | -0.1179 | No | ||
| 91 | BDNF | 10515 | 1.257 | -0.1229 | No | ||
| 92 | ADAMTS9 | 10806 | 1.186 | -0.1349 | No | ||
| 93 | HOXA10 | 11366 | 1.035 | -0.1594 | No | ||
| 94 | SP7 | 11403 | 1.022 | -0.1599 | No | ||
| 95 | ADRB1 | 11527 | 0.985 | -0.1644 | No | ||
| 96 | TENC1 | 11753 | 0.919 | -0.1737 | No | ||
| 97 | CALM2 | 11871 | 0.882 | -0.1781 | No | ||
| 98 | HOXB1 | 12024 | 0.834 | -0.1842 | No | ||
| 99 | ZBTB3 | 12060 | 0.824 | -0.1848 | No | ||
| 100 | EPHB1 | 12428 | 0.716 | -0.2009 | No | ||
| 101 | HOXC10 | 12756 | 0.605 | -0.2152 | No | ||
| 102 | KCNV2 | 12870 | 0.562 | -0.2198 | No | ||
| 103 | MLLT7 | 12965 | 0.528 | -0.2235 | No | ||
| 104 | MOV10 | 13144 | 0.459 | -0.2312 | No | ||
| 105 | BLOC1S3 | 13346 | 0.377 | -0.2400 | No | ||
| 106 | KCNK4 | 13359 | 0.372 | -0.2401 | No | ||
| 107 | HOXB8 | 14620 | -0.283 | -0.2977 | No | ||
| 108 | KCNC1 | 14867 | -0.456 | -0.3085 | No | ||
| 109 | ACCN2 | 14958 | -0.505 | -0.3121 | No | ||
| 110 | KCNN2 | 15254 | -0.711 | -0.3248 | No | ||
| 111 | MAP1A | 15548 | -0.913 | -0.3372 | No | ||
| 112 | STK35 | 16140 | -1.357 | -0.3629 | No | ||
| 113 | GRK5 | 16384 | -1.558 | -0.3723 | No | ||
| 114 | NCDN | 16895 | -2.025 | -0.3934 | No | ||
| 115 | CRMP1 | 17054 | -2.182 | -0.3982 | No | ||
| 116 | TIAM1 | 17271 | -2.359 | -0.4055 | No | ||
| 117 | XPO7 | 17391 | -2.472 | -0.4082 | No | ||
| 118 | SIGIRR | 17477 | -2.576 | -0.4092 | No | ||
| 119 | RASGEF1A | 17514 | -2.616 | -0.4079 | No | ||
| 120 | RBM12 | 17668 | -2.752 | -0.4118 | No | ||
| 121 | PAX8 | 17776 | -2.867 | -0.4135 | No | ||
| 122 | DLGAP4 | 18033 | -3.133 | -0.4218 | No | ||
| 123 | TBCC | 18075 | -3.184 | -0.4201 | No | ||
| 124 | ADNP | 18312 | -3.434 | -0.4270 | No | ||
| 125 | LMO4 | 18411 | -3.547 | -0.4275 | No | ||
| 126 | MAZ | 18575 | -3.730 | -0.4308 | No | ||
| 127 | MYO1C | 18613 | -3.770 | -0.4283 | No | ||
| 128 | F13A1 | 18705 | -3.887 | -0.4281 | No | ||
| 129 | ABHD8 | 18856 | -4.087 | -0.4304 | No | ||
| 130 | TPT1 | 19164 | -4.528 | -0.4394 | Yes | ||
| 131 | FOS | 19178 | -4.546 | -0.4349 | Yes | ||
| 132 | PCF11 | 19184 | -4.553 | -0.4300 | Yes | ||
| 133 | ZFP91 | 19257 | -4.657 | -0.4280 | Yes | ||
| 134 | PRKAG1 | 19286 | -4.710 | -0.4240 | Yes | ||
| 135 | WDTC1 | 19313 | -4.751 | -0.4199 | Yes | ||
| 136 | ZRANB1 | 19362 | -4.828 | -0.4166 | Yes | ||
| 137 | EPS15 | 19457 | -4.980 | -0.4153 | Yes | ||
| 138 | FOXJ2 | 19463 | -4.985 | -0.4100 | Yes | ||
| 139 | SP4 | 19487 | -5.016 | -0.4054 | Yes | ||
| 140 | NFKB2 | 19492 | -5.020 | -0.3999 | Yes | ||
| 141 | PPP1R10 | 19522 | -5.073 | -0.3955 | Yes | ||
| 142 | EIF4E | 19561 | -5.139 | -0.3915 | Yes | ||
| 143 | SLC25A23 | 19571 | -5.151 | -0.3861 | Yes | ||
| 144 | TOM1L2 | 19592 | -5.179 | -0.3812 | Yes | ||
| 145 | UBAP1 | 19753 | -5.409 | -0.3824 | Yes | ||
| 146 | CHD4 | 19790 | -5.476 | -0.3779 | Yes | ||
| 147 | CYLD | 19870 | -5.610 | -0.3752 | Yes | ||
| 148 | GATA3 | 20113 | -6.093 | -0.3795 | Yes | ||
| 149 | SEC24B | 20156 | -6.176 | -0.3745 | Yes | ||
| 150 | TBC1D13 | 20183 | -6.229 | -0.3687 | Yes | ||
| 151 | AKT2 | 20209 | -6.297 | -0.3627 | Yes | ||
| 152 | ARNTL | 20247 | -6.382 | -0.3572 | Yes | ||
| 153 | HIVEP1 | 20328 | -6.553 | -0.3535 | Yes | ||
| 154 | DDX17 | 20343 | -6.577 | -0.3468 | Yes | ||
| 155 | CNOT4 | 20347 | -6.588 | -0.3395 | Yes | ||
| 156 | CAMTA2 | 20353 | -6.602 | -0.3323 | Yes | ||
| 157 | LDB1 | 20425 | -6.795 | -0.3279 | Yes | ||
| 158 | AMIGO2 | 20585 | -7.160 | -0.3271 | Yes | ||
| 159 | DCTN1 | 20610 | -7.248 | -0.3201 | Yes | ||
| 160 | STK4 | 20820 | -7.881 | -0.3208 | Yes | ||
| 161 | HIF3A | 20869 | -8.066 | -0.3139 | Yes | ||
| 162 | BAZ2A | 20900 | -8.167 | -0.3061 | Yes | ||
| 163 | PRKCBP1 | 20918 | -8.231 | -0.2976 | Yes | ||
| 164 | CPNE1 | 20927 | -8.271 | -0.2887 | Yes | ||
| 165 | PITPNC1 | 20931 | -8.288 | -0.2795 | Yes | ||
| 166 | OSBPL7 | 21000 | -8.576 | -0.2730 | Yes | ||
| 167 | GNAI2 | 21032 | -8.712 | -0.2646 | Yes | ||
| 168 | FBS1 | 21041 | -8.734 | -0.2551 | Yes | ||
| 169 | MBD6 | 21121 | -9.030 | -0.2486 | Yes | ||
| 170 | SRRM1 | 21127 | -9.050 | -0.2386 | Yes | ||
| 171 | CCND2 | 21139 | -9.105 | -0.2289 | Yes | ||
| 172 | PICALM | 21330 | -10.065 | -0.2263 | Yes | ||
| 173 | NAB2 | 21372 | -10.315 | -0.2165 | Yes | ||
| 174 | DPYSL2 | 21409 | -10.591 | -0.2063 | Yes | ||
| 175 | PHF12 | 21543 | -11.631 | -0.1993 | Yes | ||
| 176 | PPP2R5C | 21618 | -12.520 | -0.1886 | Yes | ||
| 177 | CTCF | 21662 | -13.154 | -0.1757 | Yes | ||
| 178 | WHSC1L1 | 21772 | -15.077 | -0.1638 | Yes | ||
| 179 | UBE2B | 21773 | -15.079 | -0.1468 | Yes | ||
| 180 | SP1 | 21811 | -15.984 | -0.1305 | Yes | ||
| 181 | SSBP2 | 21905 | -20.594 | -0.1116 | Yes | ||
| 182 | NLK | 21913 | -21.118 | -0.0881 | Yes | ||
| 183 | FGF13 | 21947 | -79.619 | 0.0000 | Yes |