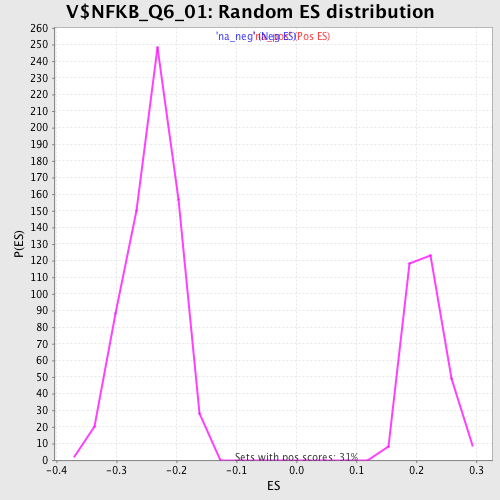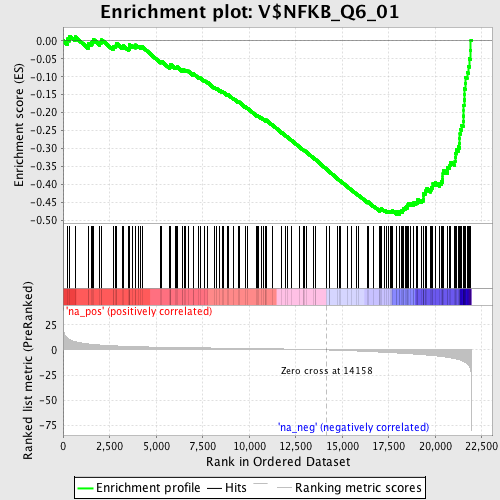
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFKB_Q6_01 |
| Enrichment Score (ES) | -0.48286697 |
| Normalized Enrichment Score (NES) | -2.0134416 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0038313558 |
| FWER p-Value | 0.0090 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF5A | 229 | 11.555 | 0.0050 | No | ||
| 2 | ARHGAP8 | 357 | 10.197 | 0.0129 | No | ||
| 3 | PCBP4 | 645 | 8.231 | 0.0107 | No | ||
| 4 | DUSP22 | 1346 | 5.915 | -0.0135 | No | ||
| 5 | GRIN2D | 1374 | 5.854 | -0.0068 | No | ||
| 6 | PCSK2 | 1505 | 5.586 | -0.0053 | No | ||
| 7 | SEMA3B | 1591 | 5.423 | -0.0019 | No | ||
| 8 | ASCL3 | 1635 | 5.340 | 0.0033 | No | ||
| 9 | ELMO1 | 1975 | 4.839 | -0.0058 | No | ||
| 10 | UBE2D3 | 2039 | 4.762 | -0.0023 | No | ||
| 11 | TGIF | 2078 | 4.723 | 0.0023 | No | ||
| 12 | EHF | 2688 | 4.028 | -0.0202 | No | ||
| 13 | EDG5 | 2714 | 4.003 | -0.0160 | No | ||
| 14 | SMPD3 | 2799 | 3.951 | -0.0146 | No | ||
| 15 | ENO3 | 2845 | 3.912 | -0.0114 | No | ||
| 16 | TP53 | 2880 | 3.885 | -0.0077 | No | ||
| 17 | VCAM1 | 3179 | 3.677 | -0.0165 | No | ||
| 18 | TRIM47 | 3232 | 3.632 | -0.0140 | No | ||
| 19 | TCEA2 | 3518 | 3.426 | -0.0224 | No | ||
| 20 | IL2RA | 3564 | 3.387 | -0.0200 | No | ||
| 21 | ZIC4 | 3571 | 3.386 | -0.0157 | No | ||
| 22 | UACA | 3572 | 3.385 | -0.0111 | No | ||
| 23 | SOX5 | 3742 | 3.284 | -0.0145 | No | ||
| 24 | DCAMKL1 | 3869 | 3.220 | -0.0160 | No | ||
| 25 | COL27A1 | 3871 | 3.220 | -0.0117 | No | ||
| 26 | NR2F2 | 4054 | 3.112 | -0.0159 | No | ||
| 27 | HSD3B7 | 4167 | 3.063 | -0.0169 | No | ||
| 28 | SP6 | 4248 | 3.020 | -0.0165 | No | ||
| 29 | CSF3 | 5239 | 2.642 | -0.0584 | No | ||
| 30 | BCL6B | 5275 | 2.630 | -0.0565 | No | ||
| 31 | IGSF4 | 5733 | 2.485 | -0.0741 | No | ||
| 32 | PTGES | 5754 | 2.476 | -0.0717 | No | ||
| 33 | HTR3B | 5761 | 2.474 | -0.0687 | No | ||
| 34 | PTPRJ | 5775 | 2.469 | -0.0660 | No | ||
| 35 | DDR1 | 6027 | 2.387 | -0.0743 | No | ||
| 36 | RIN2 | 6085 | 2.370 | -0.0737 | No | ||
| 37 | ALG6 | 6129 | 2.359 | -0.0725 | No | ||
| 38 | GREM1 | 6401 | 2.278 | -0.0819 | No | ||
| 39 | MOBKL2C | 6429 | 2.270 | -0.0801 | No | ||
| 40 | IL12A | 6521 | 2.246 | -0.0813 | No | ||
| 41 | GATA4 | 6600 | 2.225 | -0.0819 | No | ||
| 42 | EDN2 | 6712 | 2.195 | -0.0840 | No | ||
| 43 | FUT7 | 6996 | 2.120 | -0.0941 | No | ||
| 44 | PHOX2B | 7001 | 2.119 | -0.0915 | No | ||
| 45 | PTHLH | 7283 | 2.043 | -0.1016 | No | ||
| 46 | KRTAP13-1 | 7380 | 2.017 | -0.1033 | No | ||
| 47 | DGKI | 7620 | 1.957 | -0.1117 | No | ||
| 48 | WNT10A | 7750 | 1.917 | -0.1150 | No | ||
| 49 | PAPPA | 8132 | 1.831 | -0.1301 | No | ||
| 50 | CNTNAP1 | 8237 | 1.803 | -0.1324 | No | ||
| 51 | SNAP25 | 8428 | 1.761 | -0.1388 | No | ||
| 52 | SLC6A12 | 8538 | 1.734 | -0.1415 | No | ||
| 53 | FGF17 | 8615 | 1.715 | -0.1426 | No | ||
| 54 | CLCN1 | 8807 | 1.671 | -0.1492 | No | ||
| 55 | TNFSF7 | 8865 | 1.656 | -0.1496 | No | ||
| 56 | ETV1 | 9162 | 1.584 | -0.1610 | No | ||
| 57 | NFKBIB | 9422 | 1.525 | -0.1709 | No | ||
| 58 | AMOTL1 | 9451 | 1.517 | -0.1701 | No | ||
| 59 | CENTB5 | 9783 | 1.438 | -0.1834 | No | ||
| 60 | C1QL1 | 9905 | 1.408 | -0.1871 | No | ||
| 61 | KLK9 | 10415 | 1.283 | -0.2087 | No | ||
| 62 | IL7 | 10420 | 1.282 | -0.2072 | No | ||
| 63 | BDNF | 10515 | 1.257 | -0.2098 | No | ||
| 64 | NDRG2 | 10654 | 1.225 | -0.2145 | No | ||
| 65 | MSX1 | 10780 | 1.194 | -0.2186 | No | ||
| 66 | POU2F3 | 10885 | 1.165 | -0.2218 | No | ||
| 67 | TBX20 | 10895 | 1.162 | -0.2207 | No | ||
| 68 | BLR1 | 10918 | 1.155 | -0.2201 | No | ||
| 69 | TCF1 | 11247 | 1.070 | -0.2338 | No | ||
| 70 | MSC | 11757 | 0.918 | -0.2559 | No | ||
| 71 | TSLP | 11955 | 0.854 | -0.2638 | No | ||
| 72 | SHOX2 | 12047 | 0.828 | -0.2669 | No | ||
| 73 | EGF | 12295 | 0.756 | -0.2772 | No | ||
| 74 | BAI2 | 12714 | 0.624 | -0.2955 | No | ||
| 75 | NEK8 | 12915 | 0.545 | -0.3040 | No | ||
| 76 | MADCAM1 | 12934 | 0.538 | -0.3041 | No | ||
| 77 | BMP2K | 12987 | 0.517 | -0.3058 | No | ||
| 78 | RALGDS | 13052 | 0.495 | -0.3081 | No | ||
| 79 | GNG4 | 13441 | 0.342 | -0.3254 | No | ||
| 80 | IL23A | 13545 | 0.297 | -0.3298 | No | ||
| 81 | IL13 | 13554 | 0.294 | -0.3297 | No | ||
| 82 | DAP3 | 14129 | 0.012 | -0.3561 | No | ||
| 83 | EBI3 | 14287 | -0.084 | -0.3632 | No | ||
| 84 | RND1 | 14302 | -0.094 | -0.3637 | No | ||
| 85 | BATF | 14761 | -0.391 | -0.3842 | No | ||
| 86 | MAG | 14840 | -0.439 | -0.3872 | No | ||
| 87 | IFNB1 | 14917 | -0.487 | -0.3900 | No | ||
| 88 | FGF1 | 15255 | -0.712 | -0.4045 | No | ||
| 89 | UPF2 | 15475 | -0.862 | -0.4134 | No | ||
| 90 | IL1RN | 15746 | -1.044 | -0.4244 | No | ||
| 91 | IER5 | 15887 | -1.146 | -0.4293 | No | ||
| 92 | UBD | 16359 | -1.538 | -0.4489 | No | ||
| 93 | GRK5 | 16384 | -1.558 | -0.4479 | No | ||
| 94 | ZBTB11 | 16687 | -1.827 | -0.4593 | No | ||
| 95 | NR3C1 | 17008 | -2.137 | -0.4711 | No | ||
| 96 | GPHN | 17026 | -2.154 | -0.4690 | No | ||
| 97 | RASGRP4 | 17073 | -2.197 | -0.4682 | No | ||
| 98 | MITF | 17114 | -2.231 | -0.4670 | No | ||
| 99 | RBMS1 | 17267 | -2.356 | -0.4708 | No | ||
| 100 | ICAM1 | 17395 | -2.482 | -0.4733 | No | ||
| 101 | BMF | 17491 | -2.595 | -0.4742 | No | ||
| 102 | MMP9 | 17583 | -2.678 | -0.4748 | No | ||
| 103 | MAML2 | 17640 | -2.728 | -0.4737 | No | ||
| 104 | RHOG | 17692 | -2.775 | -0.4723 | No | ||
| 105 | REV3L | 17923 | -3.021 | -0.4788 | Yes | ||
| 106 | JAK3 | 17929 | -3.030 | -0.4750 | Yes | ||
| 107 | SOCS2 | 18085 | -3.196 | -0.4778 | Yes | ||
| 108 | TOP1 | 18087 | -3.196 | -0.4735 | Yes | ||
| 109 | IL4I1 | 18167 | -3.291 | -0.4728 | Yes | ||
| 110 | SLAMF8 | 18251 | -3.383 | -0.4720 | Yes | ||
| 111 | ACTN1 | 18273 | -3.406 | -0.4684 | Yes | ||
| 112 | BCL3 | 18305 | -3.430 | -0.4652 | Yes | ||
| 113 | TNFRSF1B | 18378 | -3.515 | -0.4638 | Yes | ||
| 114 | SIN3A | 18427 | -3.568 | -0.4612 | Yes | ||
| 115 | ILK | 18512 | -3.670 | -0.4602 | Yes | ||
| 116 | PRKCD | 18528 | -3.688 | -0.4559 | Yes | ||
| 117 | XPO6 | 18572 | -3.723 | -0.4529 | Yes | ||
| 118 | DUSP6 | 18680 | -3.854 | -0.4526 | Yes | ||
| 119 | SIRT2 | 18816 | -4.019 | -0.4534 | Yes | ||
| 120 | ERN1 | 18848 | -4.074 | -0.4494 | Yes | ||
| 121 | CREB1 | 18962 | -4.227 | -0.4489 | Yes | ||
| 122 | ANKHD1 | 19026 | -4.321 | -0.4460 | Yes | ||
| 123 | ATP1B1 | 19061 | -4.370 | -0.4416 | Yes | ||
| 124 | RAP2C | 19252 | -4.653 | -0.4441 | Yes | ||
| 125 | MAP3K11 | 19341 | -4.796 | -0.4417 | Yes | ||
| 126 | GNB1 | 19355 | -4.818 | -0.4358 | Yes | ||
| 127 | STAT6 | 19356 | -4.818 | -0.4294 | Yes | ||
| 128 | UBE2I | 19367 | -4.845 | -0.4233 | Yes | ||
| 129 | LTA | 19451 | -4.972 | -0.4205 | Yes | ||
| 130 | NFKB2 | 19492 | -5.020 | -0.4156 | Yes | ||
| 131 | NFAT5 | 19545 | -5.113 | -0.4111 | Yes | ||
| 132 | TRAF4 | 19752 | -5.405 | -0.4133 | Yes | ||
| 133 | CHD4 | 19790 | -5.476 | -0.4076 | Yes | ||
| 134 | CYLD | 19870 | -5.610 | -0.4037 | Yes | ||
| 135 | ARHGEF2 | 19872 | -5.614 | -0.3962 | Yes | ||
| 136 | PFN1 | 19993 | -5.863 | -0.3938 | Yes | ||
| 137 | TRIB2 | 20241 | -6.377 | -0.3966 | Yes | ||
| 138 | HIVEP1 | 20328 | -6.553 | -0.3918 | Yes | ||
| 139 | LTB | 20386 | -6.693 | -0.3854 | Yes | ||
| 140 | IL27RA | 20407 | -6.750 | -0.3773 | Yes | ||
| 141 | RELB | 20410 | -6.758 | -0.3683 | Yes | ||
| 142 | TNFRSF9 | 20436 | -6.820 | -0.3603 | Yes | ||
| 143 | AP1S2 | 20651 | -7.362 | -0.3602 | Yes | ||
| 144 | CHD6 | 20666 | -7.398 | -0.3509 | Yes | ||
| 145 | EP300 | 20781 | -7.751 | -0.3457 | Yes | ||
| 146 | RBPSUH | 20825 | -7.896 | -0.3371 | Yes | ||
| 147 | RFX5 | 21055 | -8.772 | -0.3358 | Yes | ||
| 148 | MRPS6 | 21064 | -8.809 | -0.3244 | Yes | ||
| 149 | USP52 | 21092 | -8.896 | -0.3137 | Yes | ||
| 150 | NTN1 | 21118 | -9.005 | -0.3027 | Yes | ||
| 151 | UNC84B | 21227 | -9.527 | -0.2949 | Yes | ||
| 152 | CCL5 | 21305 | -9.930 | -0.2851 | Yes | ||
| 153 | ZBTB9 | 21316 | -9.976 | -0.2721 | Yes | ||
| 154 | PPP1R13B | 21323 | -10.021 | -0.2589 | Yes | ||
| 155 | SEC61A1 | 21336 | -10.089 | -0.2459 | Yes | ||
| 156 | GADD45B | 21406 | -10.572 | -0.2349 | Yes | ||
| 157 | ASH1L | 21495 | -11.283 | -0.2238 | Yes | ||
| 158 | BAZ2B | 21508 | -11.364 | -0.2091 | Yes | ||
| 159 | BIRC3 | 21517 | -11.412 | -0.1941 | Yes | ||
| 160 | RASSF2 | 21521 | -11.453 | -0.1789 | Yes | ||
| 161 | UBE2H | 21546 | -11.654 | -0.1643 | Yes | ||
| 162 | ZNRF1 | 21565 | -11.884 | -0.1492 | Yes | ||
| 163 | SMOC1 | 21571 | -11.933 | -0.1334 | Yes | ||
| 164 | CD69 | 21606 | -12.413 | -0.1183 | Yes | ||
| 165 | FBXL12 | 21617 | -12.513 | -0.1019 | Yes | ||
| 166 | TNIP1 | 21705 | -13.849 | -0.0873 | Yes | ||
| 167 | NFKBIA | 21775 | -15.114 | -0.0702 | Yes | ||
| 168 | MAP4K2 | 21842 | -17.169 | -0.0502 | Yes | ||
| 169 | IL6ST | 21897 | -19.754 | -0.0261 | Yes | ||
| 170 | NLK | 21913 | -21.118 | 0.0016 | Yes |