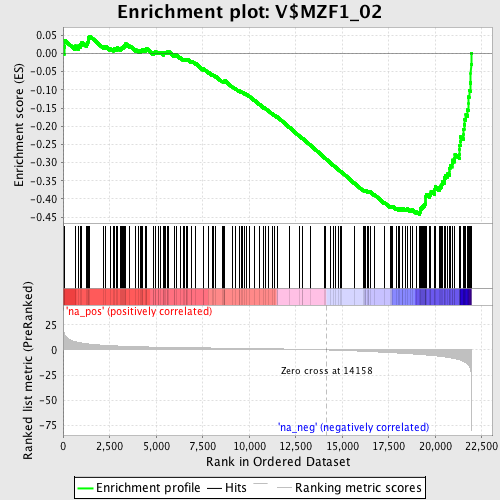
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MZF1_02 |
| Enrichment Score (ES) | -0.4427222 |
| Normalized Enrichment Score (NES) | -1.8606234 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004754223 |
| FWER p-Value | 0.074 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GART | 50 | 16.738 | 0.0184 | No | ||
| 2 | MRPS18B | 71 | 15.705 | 0.0368 | No | ||
| 3 | PCBP4 | 645 | 8.231 | 0.0207 | No | ||
| 4 | MRPL14 | 845 | 7.414 | 0.0207 | No | ||
| 5 | CGGBP1 | 922 | 7.135 | 0.0260 | No | ||
| 6 | PRRG2 | 1007 | 6.862 | 0.0306 | No | ||
| 7 | TCF2 | 1272 | 6.112 | 0.0260 | No | ||
| 8 | TFDP2 | 1321 | 5.981 | 0.0312 | No | ||
| 9 | XPO1 | 1342 | 5.926 | 0.0376 | No | ||
| 10 | GRIN2D | 1374 | 5.854 | 0.0434 | No | ||
| 11 | FGF5 | 1436 | 5.737 | 0.0477 | No | ||
| 12 | VAX1 | 2180 | 4.562 | 0.0192 | No | ||
| 13 | HIP2 | 2290 | 4.429 | 0.0196 | No | ||
| 14 | ELAVL4 | 2557 | 4.150 | 0.0125 | No | ||
| 15 | EHF | 2688 | 4.028 | 0.0115 | No | ||
| 16 | SEMA4C | 2743 | 3.985 | 0.0140 | No | ||
| 17 | TP53 | 2880 | 3.885 | 0.0125 | No | ||
| 18 | RCOR2 | 2916 | 3.860 | 0.0157 | No | ||
| 19 | PPAP2B | 3074 | 3.752 | 0.0131 | No | ||
| 20 | NTRK3 | 3133 | 3.707 | 0.0150 | No | ||
| 21 | ZNF462 | 3177 | 3.679 | 0.0176 | No | ||
| 22 | HNRPA2B1 | 3240 | 3.624 | 0.0192 | No | ||
| 23 | MRC2 | 3280 | 3.598 | 0.0218 | No | ||
| 24 | GGN | 3335 | 3.546 | 0.0237 | No | ||
| 25 | SYT7 | 3373 | 3.517 | 0.0264 | No | ||
| 26 | ORC4L | 3588 | 3.369 | 0.0207 | No | ||
| 27 | PTMA | 3909 | 3.196 | 0.0100 | No | ||
| 28 | SUPT16H | 4044 | 3.117 | 0.0077 | No | ||
| 29 | PHF15 | 4144 | 3.073 | 0.0069 | No | ||
| 30 | EYA1 | 4201 | 3.043 | 0.0081 | No | ||
| 31 | H1F0 | 4256 | 3.016 | 0.0093 | No | ||
| 32 | NFIX | 4276 | 3.007 | 0.0122 | No | ||
| 33 | GPC6 | 4432 | 2.939 | 0.0087 | No | ||
| 34 | PRDM10 | 4463 | 2.926 | 0.0109 | No | ||
| 35 | TCEB1 | 4482 | 2.920 | 0.0137 | No | ||
| 36 | ATP1A2 | 4860 | 2.771 | -0.0002 | No | ||
| 37 | SSTR3 | 4881 | 2.762 | 0.0023 | No | ||
| 38 | FOXL2 | 4954 | 2.736 | 0.0023 | No | ||
| 39 | BAT2 | 4964 | 2.733 | 0.0053 | No | ||
| 40 | ROR1 | 5110 | 2.683 | 0.0020 | No | ||
| 41 | STC2 | 5149 | 2.671 | 0.0035 | No | ||
| 42 | NLGN2 | 5235 | 2.643 | 0.0029 | No | ||
| 43 | NFATC4 | 5397 | 2.593 | -0.0013 | No | ||
| 44 | THPO | 5398 | 2.593 | 0.0019 | No | ||
| 45 | KLF12 | 5437 | 2.581 | 0.0033 | No | ||
| 46 | KCNS3 | 5499 | 2.556 | 0.0037 | No | ||
| 47 | LHX9 | 5610 | 2.523 | 0.0017 | No | ||
| 48 | NEUROD2 | 5623 | 2.521 | 0.0043 | No | ||
| 49 | LRRTM1 | 5667 | 2.505 | 0.0054 | No | ||
| 50 | AAMP | 5968 | 2.406 | -0.0054 | No | ||
| 51 | PIK3AP1 | 5991 | 2.399 | -0.0035 | No | ||
| 52 | RUNX3 | 6067 | 2.376 | -0.0040 | No | ||
| 53 | WBP4 | 6302 | 2.304 | -0.0119 | No | ||
| 54 | P4HA1 | 6450 | 2.264 | -0.0158 | No | ||
| 55 | CXCL14 | 6502 | 2.251 | -0.0154 | No | ||
| 56 | EFNA1 | 6604 | 2.223 | -0.0173 | No | ||
| 57 | LHX4 | 6620 | 2.218 | -0.0152 | No | ||
| 58 | NOS1 | 6681 | 2.202 | -0.0153 | No | ||
| 59 | HOXC4 | 6893 | 2.143 | -0.0223 | No | ||
| 60 | FBXO24 | 6921 | 2.136 | -0.0209 | No | ||
| 61 | ADAMTSL1 | 7115 | 2.085 | -0.0272 | No | ||
| 62 | DHX40 | 7523 | 1.981 | -0.0435 | No | ||
| 63 | TLX1 | 7538 | 1.977 | -0.0417 | No | ||
| 64 | CASKIN2 | 7816 | 1.902 | -0.0521 | No | ||
| 65 | ALS2CR13 | 8014 | 1.854 | -0.0588 | No | ||
| 66 | HR | 8080 | 1.842 | -0.0595 | No | ||
| 67 | SERPINB2 | 8199 | 1.814 | -0.0627 | No | ||
| 68 | SLC6A12 | 8538 | 1.734 | -0.0761 | No | ||
| 69 | ELAVL2 | 8601 | 1.718 | -0.0768 | No | ||
| 70 | UQCRH | 8656 | 1.702 | -0.0772 | No | ||
| 71 | CLDN15 | 8675 | 1.697 | -0.0759 | No | ||
| 72 | KCNIP2 | 8694 | 1.692 | -0.0747 | No | ||
| 73 | SOX30 | 9126 | 1.594 | -0.0925 | No | ||
| 74 | CHRDL1 | 9246 | 1.565 | -0.0960 | No | ||
| 75 | LIN28 | 9452 | 1.517 | -0.1036 | No | ||
| 76 | TITF1 | 9457 | 1.515 | -0.1019 | No | ||
| 77 | CBX3 | 9582 | 1.486 | -0.1058 | No | ||
| 78 | PHYHIP | 9646 | 1.474 | -0.1068 | No | ||
| 79 | USP54 | 9753 | 1.447 | -0.1099 | No | ||
| 80 | NAGS | 9849 | 1.424 | -0.1125 | No | ||
| 81 | NOL4 | 10001 | 1.381 | -0.1178 | No | ||
| 82 | GDPD2 | 10274 | 1.317 | -0.1286 | No | ||
| 83 | GABRA3 | 10547 | 1.250 | -0.1396 | No | ||
| 84 | SEMA3F | 10792 | 1.188 | -0.1493 | No | ||
| 85 | LRRTM4 | 10859 | 1.171 | -0.1509 | No | ||
| 86 | HMX1 | 11026 | 1.127 | -0.1571 | No | ||
| 87 | TCF1 | 11247 | 1.070 | -0.1659 | No | ||
| 88 | HOXA10 | 11366 | 1.035 | -0.1701 | No | ||
| 89 | CDX1 | 11504 | 0.992 | -0.1751 | No | ||
| 90 | EVI1 | 11515 | 0.989 | -0.1744 | No | ||
| 91 | LHX6 | 12160 | 0.795 | -0.2030 | No | ||
| 92 | NF1 | 12700 | 0.627 | -0.2270 | No | ||
| 93 | SALL1 | 12849 | 0.570 | -0.2331 | No | ||
| 94 | VAT1 | 12882 | 0.559 | -0.2338 | No | ||
| 95 | CLDN6 | 13297 | 0.398 | -0.2524 | No | ||
| 96 | SLC22A17 | 14039 | 0.058 | -0.2863 | No | ||
| 97 | MDS1 | 14092 | 0.033 | -0.2887 | No | ||
| 98 | OTX1 | 14353 | -0.128 | -0.3005 | No | ||
| 99 | ZADH2 | 14541 | -0.240 | -0.3088 | No | ||
| 100 | HOXB8 | 14620 | -0.283 | -0.3120 | No | ||
| 101 | ARID1A | 14795 | -0.414 | -0.3195 | No | ||
| 102 | REL | 14913 | -0.486 | -0.3242 | No | ||
| 103 | LYPLA2 | 14973 | -0.510 | -0.3263 | No | ||
| 104 | ELF4 | 15647 | -0.972 | -0.3560 | No | ||
| 105 | ZNF24 | 16124 | -1.344 | -0.3762 | No | ||
| 106 | NFYC | 16190 | -1.402 | -0.3775 | No | ||
| 107 | POLD4 | 16222 | -1.424 | -0.3772 | No | ||
| 108 | EWSR1 | 16252 | -1.451 | -0.3767 | No | ||
| 109 | CYP1A1 | 16356 | -1.535 | -0.3795 | No | ||
| 110 | GRK5 | 16384 | -1.558 | -0.3789 | No | ||
| 111 | MTA2 | 16414 | -1.585 | -0.3782 | No | ||
| 112 | PTPRF | 16493 | -1.653 | -0.3798 | No | ||
| 113 | OXCT1 | 16713 | -1.850 | -0.3876 | No | ||
| 114 | ASXL1 | 17275 | -2.364 | -0.4104 | No | ||
| 115 | OTUB1 | 17581 | -2.677 | -0.4211 | No | ||
| 116 | MAML2 | 17640 | -2.728 | -0.4204 | No | ||
| 117 | ALDH4A1 | 17697 | -2.780 | -0.4196 | No | ||
| 118 | HIC2 | 17933 | -3.035 | -0.4266 | No | ||
| 119 | TCF7L2 | 18032 | -3.133 | -0.4272 | No | ||
| 120 | TBCC | 18075 | -3.184 | -0.4252 | No | ||
| 121 | UBE4B | 18247 | -3.380 | -0.4289 | No | ||
| 122 | XPR1 | 18257 | -3.388 | -0.4252 | No | ||
| 123 | THRAP1 | 18415 | -3.552 | -0.4280 | No | ||
| 124 | ARHGAP1 | 18490 | -3.648 | -0.4269 | No | ||
| 125 | BCL9L | 18670 | -3.842 | -0.4304 | No | ||
| 126 | H3F3B | 18766 | -3.962 | -0.4298 | No | ||
| 127 | SLC25A12 | 19006 | -4.291 | -0.4355 | No | ||
| 128 | TPT1 | 19164 | -4.528 | -0.4371 | Yes | ||
| 129 | STX16 | 19183 | -4.553 | -0.4323 | Yes | ||
| 130 | PCF11 | 19184 | -4.553 | -0.4267 | Yes | ||
| 131 | RAP2C | 19252 | -4.653 | -0.4241 | Yes | ||
| 132 | HBP1 | 19318 | -4.762 | -0.4212 | Yes | ||
| 133 | PLAG1 | 19351 | -4.812 | -0.4167 | Yes | ||
| 134 | ZBTB26 | 19424 | -4.933 | -0.4139 | Yes | ||
| 135 | BAHD1 | 19481 | -5.007 | -0.4103 | Yes | ||
| 136 | TRERF1 | 19485 | -5.011 | -0.4043 | Yes | ||
| 137 | SP4 | 19487 | -5.016 | -0.3981 | Yes | ||
| 138 | TLE3 | 19490 | -5.019 | -0.3920 | Yes | ||
| 139 | PPP1R10 | 19522 | -5.073 | -0.3872 | Yes | ||
| 140 | CHCHD7 | 19703 | -5.341 | -0.3889 | Yes | ||
| 141 | TCF8 | 19761 | -5.424 | -0.3848 | Yes | ||
| 142 | SSBP3 | 19766 | -5.434 | -0.3782 | Yes | ||
| 143 | HSD11B1 | 19972 | -5.822 | -0.3805 | Yes | ||
| 144 | PPP1R12A | 19974 | -5.824 | -0.3733 | Yes | ||
| 145 | SON | 19982 | -5.838 | -0.3665 | Yes | ||
| 146 | TNPO2 | 20207 | -6.288 | -0.3690 | Yes | ||
| 147 | MAP3K7IP2 | 20281 | -6.446 | -0.3644 | Yes | ||
| 148 | CAMTA2 | 20353 | -6.602 | -0.3595 | Yes | ||
| 149 | ENSA | 20371 | -6.643 | -0.3521 | Yes | ||
| 150 | DNMT3A | 20475 | -6.913 | -0.3483 | Yes | ||
| 151 | MSI2 | 20493 | -6.952 | -0.3405 | Yes | ||
| 152 | MGEA5 | 20549 | -7.076 | -0.3343 | Yes | ||
| 153 | NCOA6 | 20675 | -7.422 | -0.3308 | Yes | ||
| 154 | RASGRP2 | 20770 | -7.709 | -0.3256 | Yes | ||
| 155 | SLC4A2 | 20772 | -7.712 | -0.3162 | Yes | ||
| 156 | LRCH4 | 20792 | -7.792 | -0.3074 | Yes | ||
| 157 | TGFB1 | 20897 | -8.163 | -0.3021 | Yes | ||
| 158 | PRKCBP1 | 20918 | -8.231 | -0.2929 | Yes | ||
| 159 | FBS1 | 21041 | -8.734 | -0.2877 | Yes | ||
| 160 | RFX5 | 21055 | -8.772 | -0.2775 | Yes | ||
| 161 | FYN | 21301 | -9.911 | -0.2765 | Yes | ||
| 162 | MLLT10 | 21310 | -9.951 | -0.2646 | Yes | ||
| 163 | ZBTB9 | 21316 | -9.976 | -0.2525 | Yes | ||
| 164 | CHD2 | 21351 | -10.205 | -0.2415 | Yes | ||
| 165 | CDC2L5 | 21362 | -10.257 | -0.2293 | Yes | ||
| 166 | ARMCX3 | 21504 | -11.347 | -0.2217 | Yes | ||
| 167 | FOXP1 | 21510 | -11.372 | -0.2079 | Yes | ||
| 168 | PHF12 | 21543 | -11.631 | -0.1951 | Yes | ||
| 169 | PAFAH1B1 | 21585 | -12.135 | -0.1820 | Yes | ||
| 170 | MLL5 | 21645 | -12.951 | -0.1687 | Yes | ||
| 171 | VASP | 21737 | -14.401 | -0.1551 | Yes | ||
| 172 | CBX6 | 21780 | -15.212 | -0.1382 | Yes | ||
| 173 | IGF1R | 21784 | -15.295 | -0.1195 | Yes | ||
| 174 | YWHAE | 21814 | -16.134 | -0.1009 | Yes | ||
| 175 | VAMP1 | 21887 | -19.080 | -0.0807 | Yes | ||
| 176 | NLK | 21913 | -21.118 | -0.0558 | Yes | ||
| 177 | BCL11B | 21916 | -21.511 | -0.0293 | Yes | ||
| 178 | EGR3 | 21930 | -24.862 | 0.0008 | Yes |
