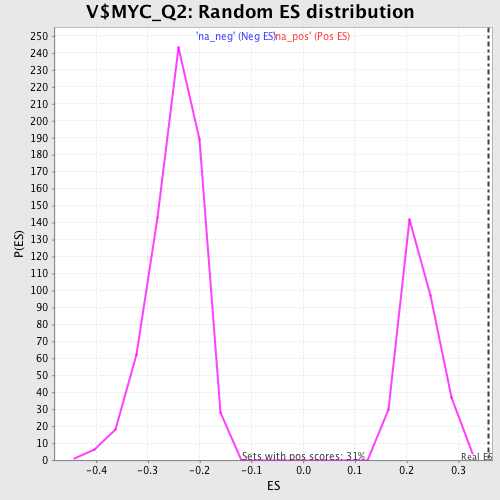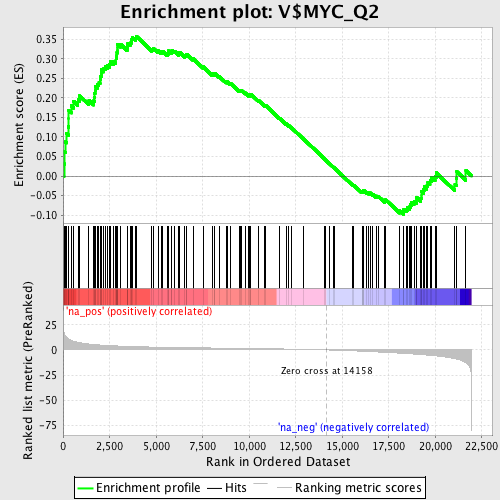
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.3581721 |
| Normalized Enrichment Score (NES) | 1.5912398 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015493085 |
| FWER p-Value | 0.53 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PA2G4 | 48 | 16.873 | 0.0310 | Yes | ||
| 2 | LDHA | 59 | 16.261 | 0.0624 | Yes | ||
| 3 | TAGLN2 | 113 | 14.192 | 0.0879 | Yes | ||
| 4 | HNRPA1 | 189 | 12.259 | 0.1086 | Yes | ||
| 5 | SHMT2 | 279 | 10.992 | 0.1261 | Yes | ||
| 6 | AMPD2 | 304 | 10.730 | 0.1461 | Yes | ||
| 7 | EBNA1BP2 | 307 | 10.700 | 0.1670 | Yes | ||
| 8 | ATAD3A | 432 | 9.491 | 0.1800 | Yes | ||
| 9 | TIMM10 | 566 | 8.600 | 0.1908 | Yes | ||
| 10 | NUDC | 799 | 7.598 | 0.1951 | Yes | ||
| 11 | NKX2-3 | 889 | 7.250 | 0.2052 | Yes | ||
| 12 | COPS7A | 1388 | 5.826 | 0.1938 | Yes | ||
| 13 | ATP5F1 | 1653 | 5.316 | 0.1922 | Yes | ||
| 14 | CTSF | 1679 | 5.278 | 0.2014 | Yes | ||
| 15 | ADK | 1692 | 5.260 | 0.2112 | Yes | ||
| 16 | AK2 | 1717 | 5.217 | 0.2204 | Yes | ||
| 17 | RPL22 | 1734 | 5.191 | 0.2298 | Yes | ||
| 18 | EIF3S10 | 1840 | 5.025 | 0.2349 | Yes | ||
| 19 | IVNS1ABP | 1927 | 4.903 | 0.2406 | Yes | ||
| 20 | AP3M1 | 1985 | 4.826 | 0.2474 | Yes | ||
| 21 | POU3F1 | 2020 | 4.780 | 0.2553 | Yes | ||
| 22 | RHEBL1 | 2050 | 4.744 | 0.2633 | Yes | ||
| 23 | FABP3 | 2062 | 4.735 | 0.2721 | Yes | ||
| 24 | POLR2L | 2189 | 4.552 | 0.2752 | Yes | ||
| 25 | COMMD3 | 2270 | 4.450 | 0.2803 | Yes | ||
| 26 | AK3 | 2380 | 4.328 | 0.2838 | Yes | ||
| 27 | ADSS | 2500 | 4.212 | 0.2866 | Yes | ||
| 28 | IPO7 | 2553 | 4.153 | 0.2924 | Yes | ||
| 29 | PAX6 | 2693 | 4.022 | 0.2940 | Yes | ||
| 30 | CTBP2 | 2839 | 3.915 | 0.2950 | Yes | ||
| 31 | B4GALT2 | 2841 | 3.914 | 0.3026 | Yes | ||
| 32 | RTN4RL2 | 2887 | 3.878 | 0.3082 | Yes | ||
| 33 | WEE1 | 2892 | 3.877 | 0.3156 | Yes | ||
| 34 | PITX3 | 2911 | 3.864 | 0.3224 | Yes | ||
| 35 | RCOR2 | 2916 | 3.860 | 0.3298 | Yes | ||
| 36 | RUSC1 | 2919 | 3.859 | 0.3373 | Yes | ||
| 37 | HMGN2 | 3078 | 3.749 | 0.3374 | Yes | ||
| 38 | SYT6 | 3438 | 3.471 | 0.3278 | Yes | ||
| 39 | FADS3 | 3470 | 3.450 | 0.3331 | Yes | ||
| 40 | FOSL1 | 3479 | 3.447 | 0.3396 | Yes | ||
| 41 | FOXD3 | 3600 | 3.362 | 0.3407 | Yes | ||
| 42 | CNNM1 | 3653 | 3.331 | 0.3448 | Yes | ||
| 43 | FGF6 | 3688 | 3.315 | 0.3498 | Yes | ||
| 44 | KRTCAP2 | 3709 | 3.305 | 0.3554 | Yes | ||
| 45 | AGMAT | 3912 | 3.194 | 0.3524 | Yes | ||
| 46 | COL2A1 | 3923 | 3.187 | 0.3582 | Yes | ||
| 47 | EIF4B | 4765 | 2.809 | 0.3251 | No | ||
| 48 | IL15RA | 4870 | 2.766 | 0.3258 | No | ||
| 49 | LTBR | 5104 | 2.685 | 0.3204 | No | ||
| 50 | PPRC1 | 5273 | 2.632 | 0.3179 | No | ||
| 51 | PSEN2 | 5339 | 2.611 | 0.3200 | No | ||
| 52 | LHX9 | 5610 | 2.523 | 0.3126 | No | ||
| 53 | CBX5 | 5641 | 2.513 | 0.3162 | No | ||
| 54 | STMN1 | 5657 | 2.509 | 0.3204 | No | ||
| 55 | SLC16A1 | 5824 | 2.456 | 0.3176 | No | ||
| 56 | GPD1 | 5846 | 2.447 | 0.3215 | No | ||
| 57 | OPRD1 | 5988 | 2.400 | 0.3197 | No | ||
| 58 | TAF6L | 6225 | 2.328 | 0.3135 | No | ||
| 59 | TDRD1 | 6245 | 2.320 | 0.3172 | No | ||
| 60 | MICAL2 | 6529 | 2.243 | 0.3086 | No | ||
| 61 | HPCA | 6613 | 2.220 | 0.3092 | No | ||
| 62 | UBXD3 | 6645 | 2.211 | 0.3121 | No | ||
| 63 | SLC1A7 | 6986 | 2.122 | 0.3007 | No | ||
| 64 | PFDN2 | 7531 | 1.979 | 0.2796 | No | ||
| 65 | ALX4 | 8006 | 1.856 | 0.2615 | No | ||
| 66 | PFKFB3 | 8112 | 1.834 | 0.2603 | No | ||
| 67 | NRIP3 | 8130 | 1.831 | 0.2632 | No | ||
| 68 | FXYD2 | 8377 | 1.775 | 0.2554 | No | ||
| 69 | APOA5 | 8755 | 1.680 | 0.2414 | No | ||
| 70 | TGFB2 | 8826 | 1.665 | 0.2415 | No | ||
| 71 | USP2 | 8988 | 1.629 | 0.2373 | No | ||
| 72 | LIN28 | 9452 | 1.517 | 0.2190 | No | ||
| 73 | LRP8 | 9504 | 1.504 | 0.2196 | No | ||
| 74 | B3GALT2 | 9584 | 1.486 | 0.2189 | No | ||
| 75 | CENTB5 | 9783 | 1.438 | 0.2127 | No | ||
| 76 | CHRM1 | 9979 | 1.387 | 0.2065 | No | ||
| 77 | ATP6V0B | 10031 | 1.377 | 0.2069 | No | ||
| 78 | TFB2M | 10069 | 1.366 | 0.2078 | No | ||
| 79 | HPCAL4 | 10090 | 1.360 | 0.2096 | No | ||
| 80 | CGN | 10491 | 1.264 | 0.1937 | No | ||
| 81 | ZZZ3 | 10839 | 1.178 | 0.1802 | No | ||
| 82 | BHLHB3 | 10863 | 1.170 | 0.1814 | No | ||
| 83 | NR0B2 | 11627 | 0.957 | 0.1483 | No | ||
| 84 | HNRPF | 11987 | 0.845 | 0.1335 | No | ||
| 85 | SFXN2 | 12123 | 0.806 | 0.1289 | No | ||
| 86 | IPO13 | 12256 | 0.766 | 0.1244 | No | ||
| 87 | ALDH3B1 | 12924 | 0.540 | 0.0948 | No | ||
| 88 | CSDA | 14022 | 0.070 | 0.0447 | No | ||
| 89 | SLC6A15 | 14093 | 0.031 | 0.0416 | No | ||
| 90 | PLA2G4A | 14318 | -0.105 | 0.0315 | No | ||
| 91 | TRIM46 | 14546 | -0.244 | 0.0216 | No | ||
| 92 | GTF2H1 | 14585 | -0.265 | 0.0203 | No | ||
| 93 | CFL1 | 15534 | -0.904 | -0.0213 | No | ||
| 94 | LMX1A | 15618 | -0.954 | -0.0233 | No | ||
| 95 | ARL3 | 16059 | -1.291 | -0.0409 | No | ||
| 96 | HOXC13 | 16077 | -1.306 | -0.0391 | No | ||
| 97 | MYCL1 | 16119 | -1.338 | -0.0384 | No | ||
| 98 | ARHGAP20 | 16127 | -1.346 | -0.0360 | No | ||
| 99 | B3GALT6 | 16303 | -1.493 | -0.0411 | No | ||
| 100 | POGK | 16416 | -1.588 | -0.0431 | No | ||
| 101 | PTPRF | 16493 | -1.653 | -0.0434 | No | ||
| 102 | FKBP11 | 16642 | -1.790 | -0.0466 | No | ||
| 103 | SCYL1 | 16826 | -1.952 | -0.0512 | No | ||
| 104 | FXYD6 | 16920 | -2.043 | -0.0514 | No | ||
| 105 | CDC14A | 17270 | -2.359 | -0.0628 | No | ||
| 106 | FCHSD2 | 17338 | -2.428 | -0.0611 | No | ||
| 107 | SIRT1 | 18076 | -3.186 | -0.0886 | No | ||
| 108 | RAB3IL1 | 18293 | -3.419 | -0.0918 | No | ||
| 109 | RRAGC | 18306 | -3.430 | -0.0856 | No | ||
| 110 | SC5DL | 18444 | -3.594 | -0.0848 | No | ||
| 111 | ILK | 18512 | -3.670 | -0.0807 | No | ||
| 112 | TIAL1 | 18634 | -3.792 | -0.0788 | No | ||
| 113 | BCL9L | 18670 | -3.842 | -0.0728 | No | ||
| 114 | FOXJ3 | 18730 | -3.918 | -0.0679 | No | ||
| 115 | ESRRA | 18857 | -4.092 | -0.0656 | No | ||
| 116 | HPS5 | 18974 | -4.235 | -0.0626 | No | ||
| 117 | NIT1 | 18981 | -4.249 | -0.0545 | No | ||
| 118 | CCDC6 | 19224 | -4.616 | -0.0565 | No | ||
| 119 | ZFP91 | 19257 | -4.657 | -0.0488 | No | ||
| 120 | TESK2 | 19259 | -4.659 | -0.0397 | No | ||
| 121 | TMEM24 | 19371 | -4.851 | -0.0353 | No | ||
| 122 | SLC43A1 | 19399 | -4.890 | -0.0269 | No | ||
| 123 | RLF | 19551 | -5.128 | -0.0238 | No | ||
| 124 | H3F3A | 19601 | -5.190 | -0.0158 | No | ||
| 125 | USP15 | 19714 | -5.353 | -0.0104 | No | ||
| 126 | CHD4 | 19790 | -5.476 | -0.0031 | No | ||
| 127 | UVRAG | 19986 | -5.842 | -0.0006 | No | ||
| 128 | ERCC6 | 20045 | -5.967 | 0.0085 | No | ||
| 129 | RFX5 | 21055 | -8.772 | -0.0205 | No | ||
| 130 | ATF7IP | 21131 | -9.063 | -0.0061 | No | ||
| 131 | NRAS | 21157 | -9.167 | 0.0107 | No | ||
| 132 | RORC | 21644 | -12.946 | 0.0139 | No |