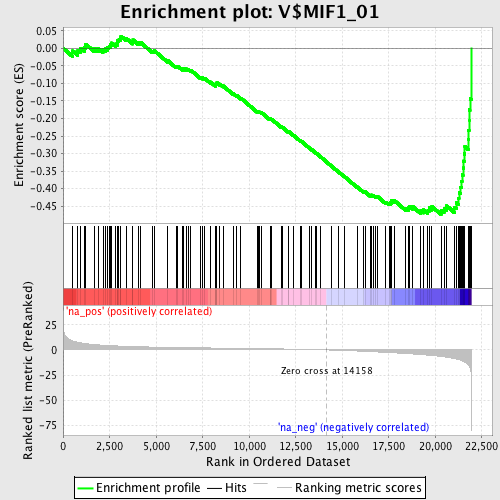
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MIF1_01 |
| Enrichment Score (ES) | -0.47372463 |
| Normalized Enrichment Score (NES) | -1.8755714 |
| Nominal p-value | 0.0014836795 |
| FDR q-value | 0.0046857195 |
| FWER p-Value | 0.062 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GRWD1 | 520 | 8.874 | -0.0074 | No | ||
| 2 | LSM7 | 794 | 7.621 | -0.0058 | No | ||
| 3 | CHCHD4 | 944 | 7.050 | 0.0004 | No | ||
| 4 | FGR | 1144 | 6.449 | 0.0032 | No | ||
| 5 | VEGF | 1209 | 6.268 | 0.0119 | No | ||
| 6 | ADK | 1692 | 5.260 | -0.0005 | No | ||
| 7 | CARS | 1906 | 4.931 | -0.0011 | No | ||
| 8 | NELF | 2145 | 4.619 | -0.0035 | No | ||
| 9 | GALNT14 | 2283 | 4.437 | -0.0015 | No | ||
| 10 | CX3CL1 | 2372 | 4.337 | 0.0025 | No | ||
| 11 | PTDSS2 | 2465 | 4.243 | 0.0061 | No | ||
| 12 | PTCH2 | 2547 | 4.156 | 0.0101 | No | ||
| 13 | WWP1 | 2601 | 4.108 | 0.0152 | No | ||
| 14 | NEK2 | 2823 | 3.930 | 0.0124 | No | ||
| 15 | TEKT2 | 2903 | 3.869 | 0.0159 | No | ||
| 16 | NAT5 | 2917 | 3.859 | 0.0225 | No | ||
| 17 | SOX3 | 3001 | 3.797 | 0.0257 | No | ||
| 18 | PLCB3 | 3072 | 3.752 | 0.0294 | No | ||
| 19 | HAT1 | 3106 | 3.728 | 0.0348 | No | ||
| 20 | MDH1B | 3383 | 3.508 | 0.0286 | No | ||
| 21 | SOX5 | 3742 | 3.284 | 0.0183 | No | ||
| 22 | TLX3 | 3754 | 3.279 | 0.0239 | No | ||
| 23 | MYCBP | 4062 | 3.110 | 0.0156 | No | ||
| 24 | PHF15 | 4144 | 3.073 | 0.0175 | No | ||
| 25 | SDCBP2 | 4814 | 2.788 | -0.0079 | No | ||
| 26 | NEDD8 | 4906 | 2.753 | -0.0070 | No | ||
| 27 | BMP6 | 5619 | 2.521 | -0.0350 | No | ||
| 28 | GRM3 | 6078 | 2.373 | -0.0515 | No | ||
| 29 | CRLF1 | 6160 | 2.349 | -0.0509 | No | ||
| 30 | MDH1 | 6423 | 2.271 | -0.0587 | No | ||
| 31 | TTC16 | 6474 | 2.260 | -0.0568 | No | ||
| 32 | SPA17 | 6608 | 2.221 | -0.0588 | No | ||
| 33 | TGFB3 | 6735 | 2.186 | -0.0605 | No | ||
| 34 | GLRA3 | 6868 | 2.148 | -0.0626 | No | ||
| 35 | PLA2G12A | 7370 | 2.019 | -0.0818 | No | ||
| 36 | GRIK5 | 7498 | 1.987 | -0.0840 | No | ||
| 37 | FIBCD1 | 7612 | 1.958 | -0.0855 | No | ||
| 38 | SMAD5 | 7917 | 1.879 | -0.0960 | No | ||
| 39 | EMX2 | 8181 | 1.818 | -0.1046 | No | ||
| 40 | KIF17 | 8191 | 1.816 | -0.1017 | No | ||
| 41 | CD44 | 8241 | 1.803 | -0.1006 | No | ||
| 42 | PACRG | 8266 | 1.797 | -0.0984 | No | ||
| 43 | THNSL1 | 8411 | 1.765 | -0.1017 | No | ||
| 44 | ELAVL2 | 8601 | 1.718 | -0.1072 | No | ||
| 45 | GRID2 | 9145 | 1.589 | -0.1291 | No | ||
| 46 | TBC1D16 | 9327 | 1.545 | -0.1345 | No | ||
| 47 | FILIP1 | 9551 | 1.493 | -0.1420 | No | ||
| 48 | PRKCSH | 10455 | 1.273 | -0.1810 | No | ||
| 49 | BDNF | 10515 | 1.257 | -0.1814 | No | ||
| 50 | DMD | 10568 | 1.245 | -0.1815 | No | ||
| 51 | MAGED1 | 10645 | 1.227 | -0.1827 | No | ||
| 52 | CPEB2 | 11118 | 1.104 | -0.2022 | No | ||
| 53 | PARK2 | 11136 | 1.100 | -0.2010 | No | ||
| 54 | STK36 | 11194 | 1.087 | -0.2016 | No | ||
| 55 | EFNB3 | 11718 | 0.928 | -0.2238 | No | ||
| 56 | MRPL10 | 11785 | 0.910 | -0.2252 | No | ||
| 57 | LRP2 | 12103 | 0.810 | -0.2382 | No | ||
| 58 | TUSC3 | 12128 | 0.803 | -0.2378 | No | ||
| 59 | STMN4 | 12372 | 0.731 | -0.2476 | No | ||
| 60 | DNAH9 | 12735 | 0.618 | -0.2630 | No | ||
| 61 | TTC8 | 12799 | 0.588 | -0.2648 | No | ||
| 62 | GPRC5D | 13264 | 0.412 | -0.2853 | No | ||
| 63 | SNF1LK | 13371 | 0.369 | -0.2895 | No | ||
| 64 | ADCK4 | 13550 | 0.295 | -0.2971 | No | ||
| 65 | RFX2 | 13639 | 0.254 | -0.3006 | No | ||
| 66 | BBS2 | 13855 | 0.146 | -0.3102 | No | ||
| 67 | DPAGT1 | 14439 | -0.178 | -0.3366 | No | ||
| 68 | ARID1A | 14795 | -0.414 | -0.3521 | No | ||
| 69 | ITGB8 | 15102 | -0.614 | -0.3650 | No | ||
| 70 | CSMD3 | 15807 | -1.084 | -0.3952 | No | ||
| 71 | SALL2 | 16153 | -1.369 | -0.4085 | No | ||
| 72 | POLD4 | 16222 | -1.424 | -0.4089 | No | ||
| 73 | WARS2 | 16505 | -1.661 | -0.4188 | No | ||
| 74 | E2F5 | 16561 | -1.721 | -0.4181 | No | ||
| 75 | NRGN | 16671 | -1.816 | -0.4197 | No | ||
| 76 | KIF3B | 16797 | -1.926 | -0.4219 | No | ||
| 77 | NCDN | 16895 | -2.025 | -0.4226 | No | ||
| 78 | TPCN1 | 17335 | -2.423 | -0.4382 | No | ||
| 79 | RASGEF1A | 17514 | -2.616 | -0.4415 | No | ||
| 80 | GTF3C4 | 17579 | -2.677 | -0.4395 | No | ||
| 81 | RNF41 | 17631 | -2.722 | -0.4368 | No | ||
| 82 | ARL4A | 17658 | -2.745 | -0.4329 | No | ||
| 83 | DDX31 | 17785 | -2.876 | -0.4334 | No | ||
| 84 | HMGCS1 | 18408 | -3.543 | -0.4553 | No | ||
| 85 | NCOA5 | 18547 | -3.706 | -0.4548 | No | ||
| 86 | MYO1C | 18613 | -3.770 | -0.4508 | No | ||
| 87 | SREBF1 | 18780 | -3.970 | -0.4511 | No | ||
| 88 | CCDC6 | 19224 | -4.616 | -0.4628 | Yes | ||
| 89 | TMEM24 | 19371 | -4.851 | -0.4605 | Yes | ||
| 90 | CBLB | 19603 | -5.196 | -0.4615 | Yes | ||
| 91 | PSCD2 | 19665 | -5.278 | -0.4545 | Yes | ||
| 92 | AP1M1 | 19815 | -5.518 | -0.4511 | Yes | ||
| 93 | HM13 | 20309 | -6.506 | -0.4617 | Yes | ||
| 94 | TBPL1 | 20470 | -6.907 | -0.4562 | Yes | ||
| 95 | DCTN1 | 20610 | -7.248 | -0.4492 | Yes | ||
| 96 | RNF25 | 21040 | -8.731 | -0.4527 | Yes | ||
| 97 | CCND2 | 21139 | -9.105 | -0.4404 | Yes | ||
| 98 | GMPR2 | 21256 | -9.640 | -0.4278 | Yes | ||
| 99 | MADD | 21274 | -9.747 | -0.4106 | Yes | ||
| 100 | SOCS1 | 21356 | -10.225 | -0.3954 | Yes | ||
| 101 | DHRS3 | 21412 | -10.613 | -0.3783 | Yes | ||
| 102 | DNAJC1 | 21437 | -10.820 | -0.3594 | Yes | ||
| 103 | TRIM39 | 21491 | -11.249 | -0.3410 | Yes | ||
| 104 | BCL2L1 | 21524 | -11.498 | -0.3212 | Yes | ||
| 105 | SPRY2 | 21549 | -11.669 | -0.3007 | Yes | ||
| 106 | DHX30 | 21588 | -12.166 | -0.2800 | Yes | ||
| 107 | CBX4 | 21769 | -14.930 | -0.2606 | Yes | ||
| 108 | CBX6 | 21780 | -15.212 | -0.2329 | Yes | ||
| 109 | YWHAE | 21814 | -16.134 | -0.2046 | Yes | ||
| 110 | NEDD9 | 21820 | -16.353 | -0.1746 | Yes | ||
| 111 | CCRK | 21865 | -17.895 | -0.1435 | Yes | ||
| 112 | FGF13 | 21947 | -79.619 | 0.0000 | Yes |