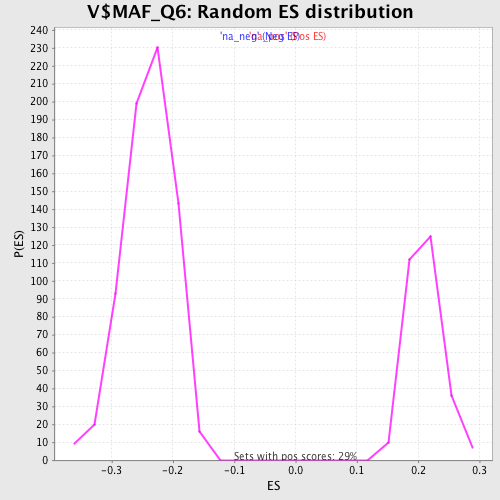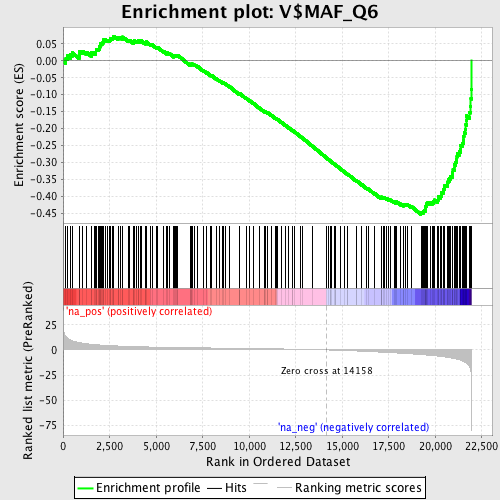
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MAF_Q6 |
| Enrichment Score (ES) | -0.45412165 |
| Normalized Enrichment Score (NES) | -1.8970557 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004133266 |
| FWER p-Value | 0.042 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KDELR2 | 152 | 13.067 | 0.0068 | No | ||
| 2 | SQLE | 226 | 11.580 | 0.0156 | No | ||
| 3 | FBXL18 | 403 | 9.788 | 0.0179 | No | ||
| 4 | CLNS1A | 488 | 9.104 | 0.0236 | No | ||
| 5 | RAB33A | 859 | 7.374 | 0.0144 | No | ||
| 6 | SYNCRIP | 870 | 7.330 | 0.0216 | No | ||
| 7 | TNFSF10 | 893 | 7.236 | 0.0283 | No | ||
| 8 | HMGA1 | 1049 | 6.736 | 0.0282 | No | ||
| 9 | TCF2 | 1272 | 6.112 | 0.0245 | No | ||
| 10 | KATNB1 | 1518 | 5.570 | 0.0191 | No | ||
| 11 | AMOT | 1537 | 5.513 | 0.0241 | No | ||
| 12 | ELOVL6 | 1672 | 5.287 | 0.0235 | No | ||
| 13 | LAMP2 | 1758 | 5.151 | 0.0250 | No | ||
| 14 | RRAS | 1767 | 5.132 | 0.0301 | No | ||
| 15 | KPNB1 | 1789 | 5.095 | 0.0345 | No | ||
| 16 | TIMELESS | 1905 | 4.932 | 0.0344 | No | ||
| 17 | CORO1C | 1947 | 4.883 | 0.0376 | No | ||
| 18 | ETV3 | 1962 | 4.857 | 0.0421 | No | ||
| 19 | SYTL1 | 1992 | 4.815 | 0.0459 | No | ||
| 20 | BAD | 2011 | 4.790 | 0.0501 | No | ||
| 21 | PEPD | 2086 | 4.714 | 0.0516 | No | ||
| 22 | EDG2 | 2136 | 4.627 | 0.0543 | No | ||
| 23 | HMGB3 | 2176 | 4.564 | 0.0573 | No | ||
| 24 | ALDOA | 2183 | 4.556 | 0.0618 | No | ||
| 25 | TYSND1 | 2259 | 4.459 | 0.0631 | No | ||
| 26 | POU2AF1 | 2407 | 4.304 | 0.0609 | No | ||
| 27 | RPS6KA3 | 2494 | 4.216 | 0.0613 | No | ||
| 28 | GAS7 | 2539 | 4.162 | 0.0637 | No | ||
| 29 | ELAVL4 | 2557 | 4.150 | 0.0673 | No | ||
| 30 | PROK2 | 2669 | 4.044 | 0.0665 | No | ||
| 31 | PAX6 | 2693 | 4.022 | 0.0696 | No | ||
| 32 | EDG5 | 2714 | 4.003 | 0.0730 | No | ||
| 33 | HOXC6 | 2951 | 3.830 | 0.0661 | No | ||
| 34 | PRDX6 | 2977 | 3.814 | 0.0690 | No | ||
| 35 | HMGN2 | 3078 | 3.749 | 0.0684 | No | ||
| 36 | VCAM1 | 3179 | 3.677 | 0.0676 | No | ||
| 37 | CDC5L | 3204 | 3.652 | 0.0704 | No | ||
| 38 | TNKS | 3495 | 3.439 | 0.0607 | No | ||
| 39 | TPBG | 3577 | 3.379 | 0.0605 | No | ||
| 40 | A2BP1 | 3775 | 3.268 | 0.0549 | No | ||
| 41 | GPC4 | 3804 | 3.253 | 0.0571 | No | ||
| 42 | CDK8 | 3816 | 3.244 | 0.0600 | No | ||
| 43 | CDH13 | 3926 | 3.186 | 0.0583 | No | ||
| 44 | SUPT16H | 4044 | 3.117 | 0.0562 | No | ||
| 45 | SIX4 | 4050 | 3.114 | 0.0593 | No | ||
| 46 | FLT1 | 4151 | 3.068 | 0.0579 | No | ||
| 47 | STAC2 | 4184 | 3.051 | 0.0597 | No | ||
| 48 | RORA | 4408 | 2.947 | 0.0525 | No | ||
| 49 | OGG1 | 4423 | 2.942 | 0.0550 | No | ||
| 50 | TWIST1 | 4465 | 2.926 | 0.0562 | No | ||
| 51 | COL1A1 | 4721 | 2.826 | 0.0474 | No | ||
| 52 | ADAM2 | 4786 | 2.798 | 0.0474 | No | ||
| 53 | SYNPR | 5007 | 2.719 | 0.0402 | No | ||
| 54 | PCDH10 | 5086 | 2.690 | 0.0394 | No | ||
| 55 | OTP | 5388 | 2.596 | 0.0283 | No | ||
| 56 | PDGFRA | 5575 | 2.533 | 0.0225 | No | ||
| 57 | RASSF3 | 5582 | 2.531 | 0.0249 | No | ||
| 58 | CUL2 | 5718 | 2.490 | 0.0213 | No | ||
| 59 | PGF | 5944 | 2.416 | 0.0135 | No | ||
| 60 | TFEB | 5962 | 2.408 | 0.0152 | No | ||
| 61 | HYAL2 | 6036 | 2.386 | 0.0144 | No | ||
| 62 | GRM3 | 6078 | 2.373 | 0.0150 | No | ||
| 63 | HOXB4 | 6138 | 2.354 | 0.0148 | No | ||
| 64 | KCNJ16 | 6155 | 2.350 | 0.0165 | No | ||
| 65 | BMP5 | 6832 | 2.159 | -0.0123 | No | ||
| 66 | CHAD | 6864 | 2.150 | -0.0114 | No | ||
| 67 | HOXC4 | 6893 | 2.143 | -0.0105 | No | ||
| 68 | ALX3 | 6916 | 2.136 | -0.0092 | No | ||
| 69 | BMP4 | 6968 | 2.126 | -0.0093 | No | ||
| 70 | VAMP3 | 7083 | 2.094 | -0.0123 | No | ||
| 71 | NPAS3 | 7220 | 2.059 | -0.0164 | No | ||
| 72 | TLX1 | 7538 | 1.977 | -0.0289 | No | ||
| 73 | GAB2 | 7726 | 1.924 | -0.0355 | No | ||
| 74 | MGLL | 7933 | 1.876 | -0.0430 | No | ||
| 75 | PGRMC2 | 7959 | 1.870 | -0.0422 | No | ||
| 76 | CNTNAP1 | 8237 | 1.803 | -0.0530 | No | ||
| 77 | SNAP25 | 8428 | 1.761 | -0.0599 | No | ||
| 78 | FXYD4 | 8584 | 1.723 | -0.0652 | No | ||
| 79 | CALN1 | 8593 | 1.721 | -0.0637 | No | ||
| 80 | BNC2 | 8700 | 1.690 | -0.0668 | No | ||
| 81 | DMP1 | 8938 | 1.641 | -0.0760 | No | ||
| 82 | LIN28 | 9452 | 1.517 | -0.0980 | No | ||
| 83 | TITF1 | 9457 | 1.515 | -0.0966 | No | ||
| 84 | NOVA2 | 9496 | 1.507 | -0.0967 | No | ||
| 85 | HAS2 | 9848 | 1.424 | -0.1113 | No | ||
| 86 | KCTD15 | 10018 | 1.378 | -0.1177 | No | ||
| 87 | INSRR | 10217 | 1.329 | -0.1254 | No | ||
| 88 | GABRA3 | 10547 | 1.250 | -0.1392 | No | ||
| 89 | CDK5 | 10811 | 1.185 | -0.1500 | No | ||
| 90 | NXPH4 | 10846 | 1.176 | -0.1503 | No | ||
| 91 | ESRRG | 10869 | 1.169 | -0.1501 | No | ||
| 92 | RASGRP3 | 10969 | 1.145 | -0.1535 | No | ||
| 93 | FGD2 | 11007 | 1.134 | -0.1540 | No | ||
| 94 | FOXP2 | 11204 | 1.084 | -0.1618 | No | ||
| 95 | SNCG | 11417 | 1.019 | -0.1705 | No | ||
| 96 | LALBA | 11489 | 0.996 | -0.1727 | No | ||
| 97 | NEGR1 | 11498 | 0.993 | -0.1720 | No | ||
| 98 | EFNB3 | 11718 | 0.928 | -0.1811 | No | ||
| 99 | CBLN2 | 11959 | 0.852 | -0.1913 | No | ||
| 100 | TUSC3 | 12128 | 0.803 | -0.1981 | No | ||
| 101 | GPX1 | 12331 | 0.745 | -0.2066 | No | ||
| 102 | SIX1 | 12349 | 0.739 | -0.2066 | No | ||
| 103 | WNT3 | 12449 | 0.708 | -0.2104 | No | ||
| 104 | SOX2 | 12771 | 0.598 | -0.2246 | No | ||
| 105 | UNC5C | 12888 | 0.556 | -0.2293 | No | ||
| 106 | PTGER2 | 13396 | 0.358 | -0.2522 | No | ||
| 107 | SLC16A6 | 14126 | 0.013 | -0.2857 | No | ||
| 108 | STX12 | 14266 | -0.071 | -0.2921 | No | ||
| 109 | OTX1 | 14353 | -0.128 | -0.2959 | No | ||
| 110 | MMRN2 | 14363 | -0.132 | -0.2962 | No | ||
| 111 | RARB | 14399 | -0.154 | -0.2976 | No | ||
| 112 | NKX2-2 | 14601 | -0.271 | -0.3066 | No | ||
| 113 | HOXB8 | 14620 | -0.283 | -0.3071 | No | ||
| 114 | MEOX2 | 14637 | -0.297 | -0.3075 | No | ||
| 115 | CNTN4 | 14887 | -0.469 | -0.3185 | No | ||
| 116 | SLC34A3 | 15110 | -0.620 | -0.3280 | No | ||
| 117 | CXXC5 | 15268 | -0.719 | -0.3345 | No | ||
| 118 | SP8 | 15289 | -0.730 | -0.3346 | No | ||
| 119 | CNOT7 | 15748 | -1.045 | -0.3546 | No | ||
| 120 | LMAN2 | 15756 | -1.052 | -0.3538 | No | ||
| 121 | PPM1E | 16058 | -1.289 | -0.3663 | No | ||
| 122 | AIF1 | 16322 | -1.506 | -0.3768 | No | ||
| 123 | ROD1 | 16398 | -1.572 | -0.3786 | No | ||
| 124 | SMARCA1 | 16742 | -1.875 | -0.3923 | No | ||
| 125 | E2F4 | 17086 | -2.203 | -0.4058 | No | ||
| 126 | BRDT | 17102 | -2.222 | -0.4041 | No | ||
| 127 | MITF | 17114 | -2.231 | -0.4023 | No | ||
| 128 | PHC1 | 17194 | -2.296 | -0.4035 | No | ||
| 129 | TIAM1 | 17271 | -2.359 | -0.4045 | No | ||
| 130 | DDX50 | 17394 | -2.481 | -0.4075 | No | ||
| 131 | BMF | 17491 | -2.595 | -0.4092 | No | ||
| 132 | NUP214 | 17596 | -2.687 | -0.4111 | No | ||
| 133 | NUMB | 17786 | -2.876 | -0.4168 | No | ||
| 134 | PTPN22 | 17874 | -2.968 | -0.4176 | No | ||
| 135 | LZTS2 | 17915 | -3.014 | -0.4163 | No | ||
| 136 | AKT3 | 18133 | -3.248 | -0.4229 | No | ||
| 137 | ACTN1 | 18273 | -3.406 | -0.4257 | No | ||
| 138 | ALCAM | 18309 | -3.432 | -0.4236 | No | ||
| 139 | HMGCS1 | 18408 | -3.543 | -0.4244 | No | ||
| 140 | SLC35B3 | 18488 | -3.644 | -0.4242 | No | ||
| 141 | ZHX2 | 18710 | -3.894 | -0.4303 | No | ||
| 142 | CD5 | 19230 | -4.624 | -0.4492 | Yes | ||
| 143 | PRKAG1 | 19286 | -4.710 | -0.4468 | Yes | ||
| 144 | STAT6 | 19356 | -4.818 | -0.4449 | Yes | ||
| 145 | RARA | 19397 | -4.888 | -0.4416 | Yes | ||
| 146 | EPS15 | 19457 | -4.980 | -0.4390 | Yes | ||
| 147 | PIK4CB | 19467 | -4.991 | -0.4342 | Yes | ||
| 148 | TRERF1 | 19485 | -5.011 | -0.4297 | Yes | ||
| 149 | CACNB3 | 19511 | -5.049 | -0.4255 | Yes | ||
| 150 | NFAT5 | 19545 | -5.113 | -0.4216 | Yes | ||
| 151 | CD79B | 19598 | -5.186 | -0.4186 | Yes | ||
| 152 | MPO | 19728 | -5.375 | -0.4188 | Yes | ||
| 153 | PNRC1 | 19828 | -5.537 | -0.4175 | Yes | ||
| 154 | CREB3L2 | 19909 | -5.677 | -0.4152 | Yes | ||
| 155 | SLC41A1 | 19955 | -5.796 | -0.4112 | Yes | ||
| 156 | TTC3 | 20122 | -6.111 | -0.4124 | Yes | ||
| 157 | SLC9A3R1 | 20149 | -6.163 | -0.4071 | Yes | ||
| 158 | AP2M1 | 20168 | -6.215 | -0.4014 | Yes | ||
| 159 | CHM | 20282 | -6.448 | -0.3998 | Yes | ||
| 160 | UTX | 20320 | -6.537 | -0.3946 | Yes | ||
| 161 | DDX17 | 20343 | -6.577 | -0.3886 | Yes | ||
| 162 | SPAG9 | 20419 | -6.779 | -0.3849 | Yes | ||
| 163 | VAV2 | 20452 | -6.862 | -0.3792 | Yes | ||
| 164 | AGPAT4 | 20495 | -6.955 | -0.3738 | Yes | ||
| 165 | SP3 | 20510 | -6.981 | -0.3671 | Yes | ||
| 166 | DYRK1A | 20638 | -7.321 | -0.3652 | Yes | ||
| 167 | AKAP13 | 20654 | -7.366 | -0.3581 | Yes | ||
| 168 | ACTR2 | 20688 | -7.465 | -0.3517 | Yes | ||
| 169 | SLC4A2 | 20772 | -7.712 | -0.3474 | Yes | ||
| 170 | STK4 | 20820 | -7.881 | -0.3413 | Yes | ||
| 171 | PIK3CD | 20906 | -8.191 | -0.3366 | Yes | ||
| 172 | CPNE1 | 20927 | -8.271 | -0.3288 | Yes | ||
| 173 | CHIC2 | 20948 | -8.353 | -0.3209 | Yes | ||
| 174 | POLG | 21020 | -8.661 | -0.3150 | Yes | ||
| 175 | PIK3CG | 21024 | -8.675 | -0.3060 | Yes | ||
| 176 | JMJD1C | 21097 | -8.917 | -0.2999 | Yes | ||
| 177 | MBD6 | 21121 | -9.030 | -0.2914 | Yes | ||
| 178 | ITPR1 | 21161 | -9.189 | -0.2835 | Yes | ||
| 179 | RNF44 | 21178 | -9.248 | -0.2745 | Yes | ||
| 180 | SLA | 21278 | -9.796 | -0.2687 | Yes | ||
| 181 | CHD2 | 21351 | -10.205 | -0.2613 | Yes | ||
| 182 | TGFBR1 | 21368 | -10.287 | -0.2512 | Yes | ||
| 183 | MAP4K5 | 21442 | -10.831 | -0.2431 | Yes | ||
| 184 | NCOA3 | 21501 | -11.314 | -0.2339 | Yes | ||
| 185 | FLOT2 | 21523 | -11.494 | -0.2227 | Yes | ||
| 186 | PHF12 | 21543 | -11.631 | -0.2113 | Yes | ||
| 187 | PPP2R5C | 21618 | -12.520 | -0.2015 | Yes | ||
| 188 | TLK2 | 21642 | -12.867 | -0.1890 | Yes | ||
| 189 | SEMA4A | 21668 | -13.174 | -0.1763 | Yes | ||
| 190 | CBX8 | 21681 | -13.369 | -0.1627 | Yes | ||
| 191 | MAP4K2 | 21842 | -17.169 | -0.1520 | Yes | ||
| 192 | ITPKB | 21894 | -19.628 | -0.1336 | Yes | ||
| 193 | NLK | 21913 | -21.118 | -0.1122 | Yes | ||
| 194 | SATB1 | 21934 | -28.181 | -0.0834 | Yes | ||
| 195 | FGF13 | 21947 | -79.619 | -0.0000 | Yes |