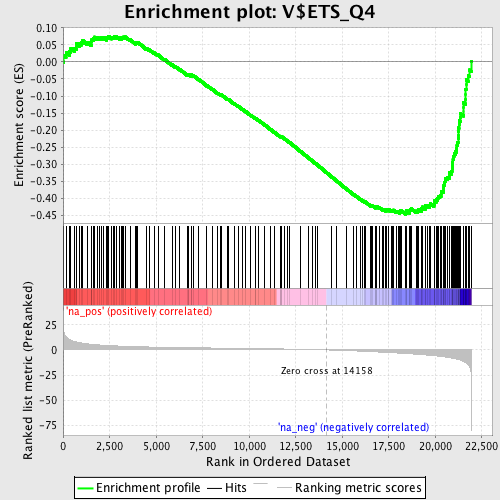
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ETS_Q4 |
| Enrichment Score (ES) | -0.44702092 |
| Normalized Enrichment Score (NES) | -1.8815805 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0045304126 |
| FWER p-Value | 0.053 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AMD1 | 32 | 18.200 | 0.0195 | No | ||
| 2 | RPS3 | 157 | 12.855 | 0.0286 | No | ||
| 3 | TCERG1 | 354 | 10.214 | 0.0314 | No | ||
| 4 | PDAP1 | 416 | 9.643 | 0.0397 | No | ||
| 5 | NASP | 620 | 8.327 | 0.0400 | No | ||
| 6 | ETF1 | 722 | 7.905 | 0.0444 | No | ||
| 7 | DPP3 | 728 | 7.875 | 0.0533 | No | ||
| 8 | NME2 | 885 | 7.275 | 0.0545 | No | ||
| 9 | SOX14 | 996 | 6.888 | 0.0574 | No | ||
| 10 | HMGA1 | 1049 | 6.736 | 0.0628 | No | ||
| 11 | E2F3 | 1330 | 5.964 | 0.0568 | No | ||
| 12 | ACSL5 | 1522 | 5.562 | 0.0544 | No | ||
| 13 | LRFN4 | 1523 | 5.559 | 0.0608 | No | ||
| 14 | RPL41 | 1547 | 5.495 | 0.0661 | No | ||
| 15 | NR1D1 | 1631 | 5.349 | 0.0684 | No | ||
| 16 | CCL2 | 1663 | 5.299 | 0.0731 | No | ||
| 17 | RNF12 | 1828 | 5.042 | 0.0714 | No | ||
| 18 | ELMO1 | 1975 | 4.839 | 0.0703 | No | ||
| 19 | LIF | 2069 | 4.729 | 0.0714 | No | ||
| 20 | RIN3 | 2166 | 4.580 | 0.0723 | No | ||
| 21 | ETV5 | 2349 | 4.365 | 0.0690 | No | ||
| 22 | EXTL2 | 2378 | 4.329 | 0.0727 | No | ||
| 23 | UBE2L3 | 2422 | 4.292 | 0.0757 | No | ||
| 24 | POU3F4 | 2608 | 4.100 | 0.0719 | No | ||
| 25 | PAX6 | 2693 | 4.022 | 0.0727 | No | ||
| 26 | SEMA4C | 2743 | 3.985 | 0.0750 | No | ||
| 27 | ARSB | 2860 | 3.899 | 0.0742 | No | ||
| 28 | RIN1 | 3026 | 3.784 | 0.0709 | No | ||
| 29 | CANX | 3142 | 3.699 | 0.0699 | No | ||
| 30 | VCAM1 | 3179 | 3.677 | 0.0725 | No | ||
| 31 | PDGFB | 3257 | 3.612 | 0.0731 | No | ||
| 32 | IRAK4 | 3342 | 3.540 | 0.0734 | No | ||
| 33 | ITPR2 | 3601 | 3.360 | 0.0654 | No | ||
| 34 | ERCC1 | 3894 | 3.203 | 0.0557 | No | ||
| 35 | UXT | 3932 | 3.184 | 0.0576 | No | ||
| 36 | NEDD4 | 4001 | 3.148 | 0.0581 | No | ||
| 37 | TWIST1 | 4465 | 2.926 | 0.0402 | No | ||
| 38 | DPPA4 | 4644 | 2.856 | 0.0354 | No | ||
| 39 | NEDD8 | 4906 | 2.753 | 0.0265 | No | ||
| 40 | LTBR | 5104 | 2.685 | 0.0206 | No | ||
| 41 | KLF12 | 5437 | 2.581 | 0.0083 | No | ||
| 42 | PPP1R14C | 5854 | 2.445 | -0.0080 | No | ||
| 43 | HYAL2 | 6036 | 2.386 | -0.0136 | No | ||
| 44 | CRSP7 | 6259 | 2.317 | -0.0211 | No | ||
| 45 | CPNE8 | 6698 | 2.197 | -0.0387 | No | ||
| 46 | TGFB3 | 6735 | 2.186 | -0.0378 | No | ||
| 47 | ADCY4 | 6749 | 2.183 | -0.0359 | No | ||
| 48 | HOXC4 | 6893 | 2.143 | -0.0400 | No | ||
| 49 | ADAMTS4 | 6897 | 2.142 | -0.0377 | No | ||
| 50 | ERG | 7011 | 2.116 | -0.0404 | No | ||
| 51 | MMP3 | 7285 | 2.043 | -0.0506 | No | ||
| 52 | GAB2 | 7726 | 1.924 | -0.0686 | No | ||
| 53 | PCDH7 | 8007 | 1.856 | -0.0793 | No | ||
| 54 | NDUFS2 | 8317 | 1.784 | -0.0914 | No | ||
| 55 | SCN3A | 8462 | 1.750 | -0.0960 | No | ||
| 56 | DRP2 | 8502 | 1.743 | -0.0958 | No | ||
| 57 | TEAD3 | 8841 | 1.662 | -0.1094 | No | ||
| 58 | CD79A | 8876 | 1.654 | -0.1091 | No | ||
| 59 | TNFSF11 | 9215 | 1.572 | -0.1228 | No | ||
| 60 | MARK1 | 9398 | 1.530 | -0.1294 | No | ||
| 61 | GJA5 | 9635 | 1.477 | -0.1385 | No | ||
| 62 | TIMP4 | 9815 | 1.430 | -0.1451 | No | ||
| 63 | GGTL3 | 10087 | 1.361 | -0.1560 | No | ||
| 64 | EPN3 | 10315 | 1.307 | -0.1649 | No | ||
| 65 | MGAT4A | 10353 | 1.298 | -0.1651 | No | ||
| 66 | RHOV | 10498 | 1.263 | -0.1703 | No | ||
| 67 | LCP1 | 10823 | 1.181 | -0.1838 | No | ||
| 68 | STARD13 | 11150 | 1.096 | -0.1975 | No | ||
| 69 | HOXA10 | 11366 | 1.035 | -0.2062 | No | ||
| 70 | SPIB | 11658 | 0.948 | -0.2185 | No | ||
| 71 | BLNK | 11697 | 0.938 | -0.2191 | No | ||
| 72 | SLC30A7 | 11745 | 0.920 | -0.2202 | No | ||
| 73 | TENC1 | 11753 | 0.919 | -0.2195 | No | ||
| 74 | CALM2 | 11871 | 0.882 | -0.2239 | No | ||
| 75 | PPP1R9B | 12037 | 0.830 | -0.2305 | No | ||
| 76 | DAB2IP | 12167 | 0.792 | -0.2355 | No | ||
| 77 | SOX2 | 12771 | 0.598 | -0.2625 | No | ||
| 78 | NDUFA4 | 13200 | 0.434 | -0.2817 | No | ||
| 79 | CSAD | 13384 | 0.363 | -0.2897 | No | ||
| 80 | IL13 | 13554 | 0.294 | -0.2971 | No | ||
| 81 | ZNF35 | 13655 | 0.243 | -0.3014 | No | ||
| 82 | POF1B | 14401 | -0.155 | -0.3354 | No | ||
| 83 | HCST | 14688 | -0.332 | -0.3482 | No | ||
| 84 | FXYD5 | 15239 | -0.704 | -0.3726 | No | ||
| 85 | LPXN | 15617 | -0.954 | -0.3889 | No | ||
| 86 | TBN | 15739 | -1.039 | -0.3932 | No | ||
| 87 | HOXA1 | 15955 | -1.200 | -0.4017 | No | ||
| 88 | BIN3 | 16091 | -1.317 | -0.4064 | No | ||
| 89 | DDX23 | 16204 | -1.409 | -0.4099 | No | ||
| 90 | POLD4 | 16222 | -1.424 | -0.4091 | No | ||
| 91 | ARHGAP15 | 16522 | -1.675 | -0.4209 | No | ||
| 92 | E2F5 | 16561 | -1.721 | -0.4206 | No | ||
| 93 | ACTR3 | 16633 | -1.785 | -0.4218 | No | ||
| 94 | CRADD | 16775 | -1.909 | -0.4261 | No | ||
| 95 | KIF3B | 16797 | -1.926 | -0.4248 | No | ||
| 96 | GPR150 | 16818 | -1.945 | -0.4235 | No | ||
| 97 | ARPC1B | 16862 | -1.997 | -0.4232 | No | ||
| 98 | HCLS1 | 17013 | -2.142 | -0.4276 | No | ||
| 99 | ACSL4 | 17183 | -2.290 | -0.4327 | No | ||
| 100 | AGPAT1 | 17240 | -2.335 | -0.4326 | No | ||
| 101 | AP1G2 | 17340 | -2.429 | -0.4344 | No | ||
| 102 | CD2BP2 | 17361 | -2.444 | -0.4325 | No | ||
| 103 | ST7L | 17458 | -2.544 | -0.4340 | No | ||
| 104 | SIGIRR | 17477 | -2.576 | -0.4318 | No | ||
| 105 | ITGB7 | 17644 | -2.731 | -0.4363 | No | ||
| 106 | FURIN | 17682 | -2.762 | -0.4348 | No | ||
| 107 | FGFR2 | 17733 | -2.815 | -0.4339 | No | ||
| 108 | CENTD2 | 17890 | -2.987 | -0.4376 | No | ||
| 109 | CENTG1 | 18001 | -3.104 | -0.4391 | No | ||
| 110 | KCNAB2 | 18098 | -3.213 | -0.4398 | No | ||
| 111 | RGS3 | 18102 | -3.221 | -0.4362 | No | ||
| 112 | SRPR | 18189 | -3.322 | -0.4363 | No | ||
| 113 | PDLIM2 | 18423 | -3.564 | -0.4429 | Yes | ||
| 114 | CAPZA1 | 18447 | -3.596 | -0.4398 | Yes | ||
| 115 | STRBP | 18459 | -3.611 | -0.4362 | Yes | ||
| 116 | BAG4 | 18610 | -3.769 | -0.4387 | Yes | ||
| 117 | PLCB2 | 18630 | -3.789 | -0.4352 | Yes | ||
| 118 | FCHO1 | 18654 | -3.816 | -0.4319 | Yes | ||
| 119 | PTPN11 | 18707 | -3.890 | -0.4298 | Yes | ||
| 120 | SEPT1 | 18972 | -4.234 | -0.4370 | Yes | ||
| 121 | ARRB2 | 19063 | -4.371 | -0.4361 | Yes | ||
| 122 | CAST | 19103 | -4.424 | -0.4328 | Yes | ||
| 123 | RAP2C | 19252 | -4.653 | -0.4343 | Yes | ||
| 124 | ZFP91 | 19257 | -4.657 | -0.4291 | Yes | ||
| 125 | PRKAG1 | 19286 | -4.710 | -0.4249 | Yes | ||
| 126 | DDIT3 | 19452 | -4.972 | -0.4268 | Yes | ||
| 127 | FOXO3A | 19458 | -4.980 | -0.4213 | Yes | ||
| 128 | DGKZ | 19562 | -5.140 | -0.4201 | Yes | ||
| 129 | DOCK11 | 19697 | -5.335 | -0.4201 | Yes | ||
| 130 | IKBKB | 19718 | -5.359 | -0.4148 | Yes | ||
| 131 | RBMS2 | 19930 | -5.732 | -0.4179 | Yes | ||
| 132 | SLC41A1 | 19955 | -5.796 | -0.4123 | Yes | ||
| 133 | SEC24C | 19971 | -5.820 | -0.4063 | Yes | ||
| 134 | VAV1 | 20066 | -6.018 | -0.4037 | Yes | ||
| 135 | GATA3 | 20113 | -6.093 | -0.3988 | Yes | ||
| 136 | SLC9A3R1 | 20149 | -6.163 | -0.3933 | Yes | ||
| 137 | CNOT10 | 20252 | -6.399 | -0.3906 | Yes | ||
| 138 | HM13 | 20309 | -6.506 | -0.3857 | Yes | ||
| 139 | UTX | 20320 | -6.537 | -0.3786 | Yes | ||
| 140 | CHES1 | 20432 | -6.813 | -0.3759 | Yes | ||
| 141 | SLC9A9 | 20442 | -6.835 | -0.3684 | Yes | ||
| 142 | USP3 | 20458 | -6.887 | -0.3612 | Yes | ||
| 143 | ELK3 | 20479 | -6.923 | -0.3541 | Yes | ||
| 144 | PIK3R4 | 20519 | -7.004 | -0.3478 | Yes | ||
| 145 | SOST | 20557 | -7.092 | -0.3413 | Yes | ||
| 146 | PTK2 | 20652 | -7.365 | -0.3372 | Yes | ||
| 147 | PML | 20746 | -7.609 | -0.3327 | Yes | ||
| 148 | CAP1 | 20757 | -7.665 | -0.3243 | Yes | ||
| 149 | LSM1 | 20858 | -8.013 | -0.3197 | Yes | ||
| 150 | BAZ2A | 20900 | -8.167 | -0.3121 | Yes | ||
| 151 | PRKCBP1 | 20918 | -8.231 | -0.3034 | Yes | ||
| 152 | RBM4 | 20935 | -8.316 | -0.2946 | Yes | ||
| 153 | CHIC2 | 20948 | -8.353 | -0.2855 | Yes | ||
| 154 | TPP2 | 20979 | -8.495 | -0.2771 | Yes | ||
| 155 | RAE1 | 21007 | -8.593 | -0.2684 | Yes | ||
| 156 | ARHGAP4 | 21091 | -8.891 | -0.2620 | Yes | ||
| 157 | GMFG | 21143 | -9.118 | -0.2538 | Yes | ||
| 158 | ITPR1 | 21161 | -9.189 | -0.2440 | Yes | ||
| 159 | GFI1 | 21194 | -9.328 | -0.2348 | Yes | ||
| 160 | CENTB1 | 21226 | -9.517 | -0.2252 | Yes | ||
| 161 | TREML2 | 21237 | -9.571 | -0.2146 | Yes | ||
| 162 | DMTF1 | 21245 | -9.609 | -0.2039 | Yes | ||
| 163 | GMPR2 | 21256 | -9.640 | -0.1932 | Yes | ||
| 164 | MADD | 21274 | -9.747 | -0.1828 | Yes | ||
| 165 | JUNB | 21298 | -9.897 | -0.1724 | Yes | ||
| 166 | SEC61A1 | 21336 | -10.089 | -0.1625 | Yes | ||
| 167 | CHD2 | 21351 | -10.205 | -0.1514 | Yes | ||
| 168 | TRIM41 | 21509 | -11.371 | -0.1455 | Yes | ||
| 169 | FOXP1 | 21510 | -11.372 | -0.1324 | Yes | ||
| 170 | PPP1R16B | 21514 | -11.386 | -0.1194 | Yes | ||
| 171 | KLF13 | 21620 | -12.575 | -0.1098 | Yes | ||
| 172 | TCF12 | 21627 | -12.637 | -0.0955 | Yes | ||
| 173 | CORO1A | 21631 | -12.679 | -0.0810 | Yes | ||
| 174 | STAT5B | 21655 | -13.076 | -0.0670 | Yes | ||
| 175 | RGS14 | 21660 | -13.135 | -0.0520 | Yes | ||
| 176 | LCP2 | 21764 | -14.815 | -0.0397 | Yes | ||
| 177 | MAP4K2 | 21842 | -17.169 | -0.0234 | Yes | ||
| 178 | DGKA | 21928 | -24.490 | 0.0009 | Yes |