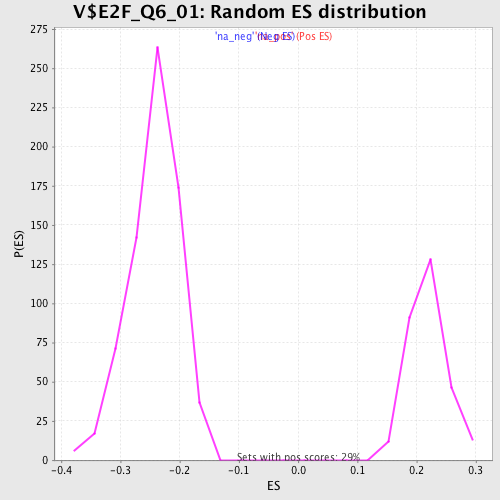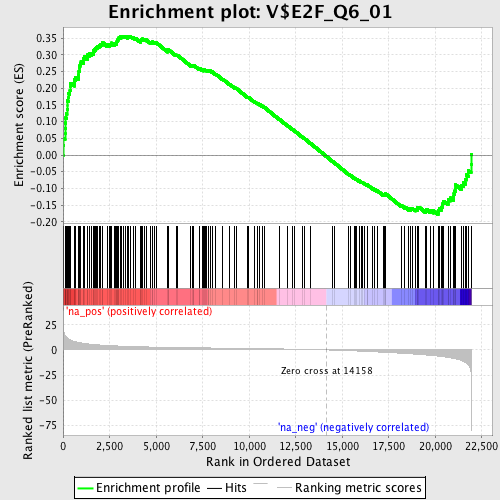
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_Q6_01 |
| Enrichment Score (ES) | 0.3560893 |
| Normalized Enrichment Score (NES) | 1.6322184 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015795143 |
| FWER p-Value | 0.389 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BMP7 | 6 | 24.610 | 0.0287 | Yes | ||
| 2 | MCM6 | 26 | 19.246 | 0.0504 | Yes | ||
| 3 | PRIM1 | 103 | 14.426 | 0.0639 | Yes | ||
| 4 | RANBP1 | 110 | 14.273 | 0.0804 | Yes | ||
| 5 | NCL | 125 | 13.890 | 0.0961 | Yes | ||
| 6 | YWHAQ | 140 | 13.364 | 0.1111 | Yes | ||
| 7 | HNRPA1 | 189 | 12.259 | 0.1234 | Yes | ||
| 8 | GMNN | 209 | 11.840 | 0.1364 | Yes | ||
| 9 | EIF5A | 229 | 11.555 | 0.1491 | Yes | ||
| 10 | STAG1 | 231 | 11.525 | 0.1626 | Yes | ||
| 11 | RBL1 | 284 | 10.907 | 0.1731 | Yes | ||
| 12 | MCM4 | 293 | 10.788 | 0.1854 | Yes | ||
| 13 | CYR61 | 367 | 10.135 | 0.1939 | Yes | ||
| 14 | MCM7 | 398 | 9.851 | 0.2041 | Yes | ||
| 15 | ACBD6 | 406 | 9.712 | 0.2152 | Yes | ||
| 16 | SFRS2 | 597 | 8.452 | 0.2164 | Yes | ||
| 17 | NASP | 620 | 8.327 | 0.2252 | Yes | ||
| 18 | TOPBP1 | 674 | 8.102 | 0.2323 | Yes | ||
| 19 | LIG1 | 820 | 7.498 | 0.2345 | Yes | ||
| 20 | DNAJC11 | 841 | 7.427 | 0.2423 | Yes | ||
| 21 | NDUFA11 | 848 | 7.395 | 0.2507 | Yes | ||
| 22 | MYC | 869 | 7.330 | 0.2584 | Yes | ||
| 23 | RFC1 | 874 | 7.311 | 0.2668 | Yes | ||
| 24 | RPS19 | 910 | 7.177 | 0.2737 | Yes | ||
| 25 | CDC45L | 960 | 6.993 | 0.2796 | Yes | ||
| 26 | UNG | 1088 | 6.592 | 0.2815 | Yes | ||
| 27 | PEG3 | 1091 | 6.573 | 0.2892 | Yes | ||
| 28 | AP4M1 | 1145 | 6.445 | 0.2943 | Yes | ||
| 29 | PLK4 | 1306 | 6.023 | 0.2941 | Yes | ||
| 30 | E2F3 | 1330 | 5.964 | 0.3000 | Yes | ||
| 31 | E2F1 | 1414 | 5.788 | 0.3030 | Yes | ||
| 32 | NUTF2 | 1511 | 5.580 | 0.3052 | Yes | ||
| 33 | FANCL | 1624 | 5.359 | 0.3063 | Yes | ||
| 34 | CDCA7 | 1636 | 5.336 | 0.3121 | Yes | ||
| 35 | RAD51 | 1676 | 5.285 | 0.3165 | Yes | ||
| 36 | PCSK4 | 1748 | 5.169 | 0.3193 | Yes | ||
| 37 | KPNB1 | 1789 | 5.095 | 0.3235 | Yes | ||
| 38 | MCM3 | 1864 | 4.991 | 0.3259 | Yes | ||
| 39 | CDC6 | 1934 | 4.895 | 0.3285 | Yes | ||
| 40 | EZH2 | 2009 | 4.793 | 0.3308 | Yes | ||
| 41 | ORC1L | 2095 | 4.703 | 0.3324 | Yes | ||
| 42 | UFD1L | 2122 | 4.650 | 0.3367 | Yes | ||
| 43 | CDC25A | 2359 | 4.348 | 0.3309 | Yes | ||
| 44 | PCNA | 2502 | 4.211 | 0.3294 | Yes | ||
| 45 | IPO7 | 2553 | 4.153 | 0.3320 | Yes | ||
| 46 | PKMYT1 | 2581 | 4.124 | 0.3356 | Yes | ||
| 47 | POLD1 | 2748 | 3.984 | 0.3326 | Yes | ||
| 48 | TMEM4 | 2815 | 3.939 | 0.3342 | Yes | ||
| 49 | XPO5 | 2886 | 3.880 | 0.3356 | Yes | ||
| 50 | WEE1 | 2892 | 3.877 | 0.3399 | Yes | ||
| 51 | RCOR2 | 2916 | 3.860 | 0.3434 | Yes | ||
| 52 | CDKN1A | 2944 | 3.835 | 0.3467 | Yes | ||
| 53 | SUV39H1 | 2996 | 3.804 | 0.3488 | Yes | ||
| 54 | PVRL1 | 3000 | 3.798 | 0.3531 | Yes | ||
| 55 | HMGN2 | 3078 | 3.749 | 0.3540 | Yes | ||
| 56 | MCM2 | 3157 | 3.689 | 0.3548 | Yes | ||
| 57 | CTNND2 | 3222 | 3.638 | 0.3561 | Yes | ||
| 58 | POLE2 | 3339 | 3.542 | 0.3549 | No | ||
| 59 | DNMT1 | 3447 | 3.462 | 0.3541 | No | ||
| 60 | MELK | 3493 | 3.439 | 0.3561 | No | ||
| 61 | SFRS1 | 3608 | 3.355 | 0.3548 | No | ||
| 62 | ANP32E | 3761 | 3.275 | 0.3516 | No | ||
| 63 | PTMA | 3909 | 3.196 | 0.3487 | No | ||
| 64 | PHF15 | 4144 | 3.073 | 0.3415 | No | ||
| 65 | KLHDC3 | 4162 | 3.064 | 0.3443 | No | ||
| 66 | ATAD2 | 4214 | 3.035 | 0.3456 | No | ||
| 67 | PAQR4 | 4246 | 3.021 | 0.3477 | No | ||
| 68 | BARHL1 | 4364 | 2.967 | 0.3458 | No | ||
| 69 | HTF9C | 4461 | 2.927 | 0.3448 | No | ||
| 70 | NRP2 | 4700 | 2.836 | 0.3373 | No | ||
| 71 | KCND2 | 4780 | 2.799 | 0.3369 | No | ||
| 72 | REPS2 | 4810 | 2.789 | 0.3389 | No | ||
| 73 | SFMBT1 | 4893 | 2.759 | 0.3383 | No | ||
| 74 | DCLRE1A | 4990 | 2.726 | 0.3371 | No | ||
| 75 | SNCB | 5618 | 2.522 | 0.3113 | No | ||
| 76 | CBX5 | 5641 | 2.513 | 0.3133 | No | ||
| 77 | STMN1 | 5657 | 2.509 | 0.3155 | No | ||
| 78 | CITED2 | 6065 | 2.376 | 0.2996 | No | ||
| 79 | RRM2 | 6132 | 2.358 | 0.2994 | No | ||
| 80 | GLRA3 | 6868 | 2.148 | 0.2681 | No | ||
| 81 | MSI1 | 6967 | 2.127 | 0.2661 | No | ||
| 82 | DMRT1 | 6985 | 2.123 | 0.2679 | No | ||
| 83 | ERG | 7011 | 2.116 | 0.2692 | No | ||
| 84 | TBX3 | 7307 | 2.036 | 0.2580 | No | ||
| 85 | RPS6KA5 | 7337 | 2.027 | 0.2591 | No | ||
| 86 | STAG2 | 7493 | 1.988 | 0.2543 | No | ||
| 87 | ADAMTS2 | 7554 | 1.973 | 0.2539 | No | ||
| 88 | CACNA1G | 7575 | 1.969 | 0.2553 | No | ||
| 89 | GRIA4 | 7645 | 1.949 | 0.2544 | No | ||
| 90 | SLITRK3 | 7711 | 1.928 | 0.2537 | No | ||
| 91 | MAT2A | 7787 | 1.908 | 0.2525 | No | ||
| 92 | ETV4 | 7815 | 1.902 | 0.2535 | No | ||
| 93 | HOXA9 | 7895 | 1.884 | 0.2521 | No | ||
| 94 | PCDH7 | 8007 | 1.856 | 0.2492 | No | ||
| 95 | SEMA5A | 8200 | 1.813 | 0.2425 | No | ||
| 96 | SLITRK4 | 8583 | 1.723 | 0.2270 | No | ||
| 97 | ARHGAP11A | 8940 | 1.640 | 0.2125 | No | ||
| 98 | BAI1 | 9222 | 1.571 | 0.2015 | No | ||
| 99 | ZIM2 | 9293 | 1.553 | 0.2001 | No | ||
| 100 | HTR2C | 9919 | 1.407 | 0.1731 | No | ||
| 101 | HNRPD | 9966 | 1.392 | 0.1726 | No | ||
| 102 | RALY | 10271 | 1.318 | 0.1602 | No | ||
| 103 | SLC6A4 | 10434 | 1.279 | 0.1542 | No | ||
| 104 | PRKCSH | 10455 | 1.273 | 0.1548 | No | ||
| 105 | DMD | 10568 | 1.245 | 0.1511 | No | ||
| 106 | UCHL1 | 10692 | 1.216 | 0.1469 | No | ||
| 107 | ADAMTS9 | 10806 | 1.186 | 0.1431 | No | ||
| 108 | SLC16A2 | 11633 | 0.956 | 0.1063 | No | ||
| 109 | PPP1R9B | 12037 | 0.830 | 0.0888 | No | ||
| 110 | KCNA6 | 12307 | 0.751 | 0.0773 | No | ||
| 111 | EPHB1 | 12428 | 0.716 | 0.0727 | No | ||
| 112 | SALL1 | 12849 | 0.570 | 0.0540 | No | ||
| 113 | FBXO5 | 12981 | 0.520 | 0.0486 | No | ||
| 114 | PODN | 13271 | 0.409 | 0.0358 | No | ||
| 115 | FANCG | 14489 | -0.216 | -0.0198 | No | ||
| 116 | NKX2-2 | 14601 | -0.271 | -0.0246 | No | ||
| 117 | CASP8AP2 | 15356 | -0.775 | -0.0583 | No | ||
| 118 | MXD3 | 15419 | -0.824 | -0.0601 | No | ||
| 119 | DLL4 | 15634 | -0.962 | -0.0688 | No | ||
| 120 | CLPTM1 | 15714 | -1.022 | -0.0713 | No | ||
| 121 | KLF5 | 15788 | -1.070 | -0.0734 | No | ||
| 122 | DDB2 | 15906 | -1.159 | -0.0774 | No | ||
| 123 | TMEM15 | 16035 | -1.268 | -0.0818 | No | ||
| 124 | FMO4 | 16061 | -1.291 | -0.0814 | No | ||
| 125 | APRIN | 16177 | -1.392 | -0.0850 | No | ||
| 126 | DAXX | 16329 | -1.510 | -0.0902 | No | ||
| 127 | PRKDC | 16339 | -1.517 | -0.0888 | No | ||
| 128 | BRMS1L | 16635 | -1.787 | -0.1003 | No | ||
| 129 | DNAJC5G | 16745 | -1.879 | -0.1031 | No | ||
| 130 | POLH | 16884 | -2.017 | -0.1070 | No | ||
| 131 | CASP2 | 17228 | -2.323 | -0.1200 | No | ||
| 132 | TIAM1 | 17271 | -2.359 | -0.1192 | No | ||
| 133 | SFRS12 | 17306 | -2.399 | -0.1179 | No | ||
| 134 | GPRC5B | 17315 | -2.402 | -0.1155 | No | ||
| 135 | ARID4A | 18192 | -3.324 | -0.1518 | No | ||
| 136 | ACIN1 | 18367 | -3.498 | -0.1557 | No | ||
| 137 | MAZ | 18575 | -3.730 | -0.1608 | No | ||
| 138 | DUSP6 | 18680 | -3.854 | -0.1610 | No | ||
| 139 | FBXL20 | 18750 | -3.942 | -0.1596 | No | ||
| 140 | BCOR | 18929 | -4.185 | -0.1628 | No | ||
| 141 | ANKHD1 | 19026 | -4.321 | -0.1621 | No | ||
| 142 | E2F7 | 19032 | -4.324 | -0.1573 | No | ||
| 143 | ATE1 | 19113 | -4.445 | -0.1557 | No | ||
| 144 | TLE3 | 19490 | -5.019 | -0.1671 | No | ||
| 145 | H2AFZ | 19510 | -5.048 | -0.1620 | No | ||
| 146 | NHLRC2 | 19760 | -5.421 | -0.1671 | No | ||
| 147 | ZCCHC8 | 19907 | -5.676 | -0.1671 | No | ||
| 148 | PPM1D | 20148 | -6.160 | -0.1709 | No | ||
| 149 | SORBS1 | 20177 | -6.222 | -0.1649 | No | ||
| 150 | TNPO2 | 20207 | -6.288 | -0.1588 | No | ||
| 151 | DDX17 | 20343 | -6.577 | -0.1573 | No | ||
| 152 | DLST | 20369 | -6.634 | -0.1506 | No | ||
| 153 | ERBB2IP | 20406 | -6.743 | -0.1444 | No | ||
| 154 | MAP3K13 | 20449 | -6.853 | -0.1382 | No | ||
| 155 | RBBP4 | 20687 | -7.464 | -0.1403 | No | ||
| 156 | RAB11B | 20694 | -7.474 | -0.1318 | No | ||
| 157 | RBPSUH | 20825 | -7.896 | -0.1285 | No | ||
| 158 | DLEU2 | 20974 | -8.472 | -0.1253 | No | ||
| 159 | OSBPL7 | 21000 | -8.576 | -0.1164 | No | ||
| 160 | ID3 | 21036 | -8.725 | -0.1078 | No | ||
| 161 | FLI1 | 21086 | -8.875 | -0.0996 | No | ||
| 162 | USP52 | 21092 | -8.896 | -0.0893 | No | ||
| 163 | DPYSL2 | 21409 | -10.591 | -0.0914 | No | ||
| 164 | LUC7L2 | 21506 | -11.357 | -0.0825 | No | ||
| 165 | AXUD1 | 21611 | -12.443 | -0.0726 | No | ||
| 166 | CTCF | 21662 | -13.154 | -0.0594 | No | ||
| 167 | WHSC1L1 | 21772 | -15.077 | -0.0467 | No | ||
| 168 | HRBL | 21919 | -21.874 | -0.0277 | No | ||
| 169 | DCK | 21929 | -24.615 | 0.0008 | No |