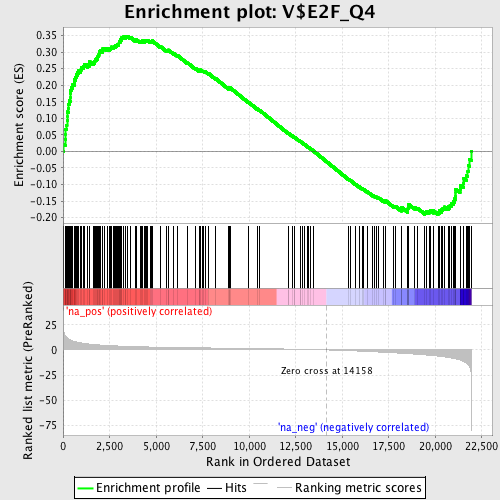
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_Q4 |
| Enrichment Score (ES) | 0.3483099 |
| Normalized Enrichment Score (NES) | 1.5984653 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015293852 |
| FWER p-Value | 0.503 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MCM6 | 26 | 19.246 | 0.0218 | Yes | ||
| 2 | RANBP1 | 110 | 14.273 | 0.0350 | Yes | ||
| 3 | SFRS7 | 119 | 13.984 | 0.0513 | Yes | ||
| 4 | NCL | 125 | 13.890 | 0.0677 | Yes | ||
| 5 | HNRPA1 | 189 | 12.259 | 0.0794 | Yes | ||
| 6 | GMNN | 209 | 11.840 | 0.0927 | Yes | ||
| 7 | NOLC1 | 225 | 11.590 | 0.1059 | Yes | ||
| 8 | STAG1 | 231 | 11.525 | 0.1194 | Yes | ||
| 9 | RBL1 | 284 | 10.907 | 0.1300 | Yes | ||
| 10 | MCM4 | 293 | 10.788 | 0.1425 | Yes | ||
| 11 | RQCD1 | 333 | 10.476 | 0.1533 | Yes | ||
| 12 | DNAJC9 | 391 | 9.910 | 0.1625 | Yes | ||
| 13 | MCM7 | 398 | 9.851 | 0.1740 | Yes | ||
| 14 | ACBD6 | 406 | 9.712 | 0.1852 | Yes | ||
| 15 | SNRPD1 | 448 | 9.347 | 0.1945 | Yes | ||
| 16 | MRPL40 | 524 | 8.847 | 0.2016 | Yes | ||
| 17 | SFRS2 | 597 | 8.452 | 0.2084 | Yes | ||
| 18 | NASP | 620 | 8.327 | 0.2173 | Yes | ||
| 19 | TOPBP1 | 674 | 8.102 | 0.2246 | Yes | ||
| 20 | CRK | 726 | 7.882 | 0.2317 | Yes | ||
| 21 | PRPS1 | 761 | 7.714 | 0.2393 | Yes | ||
| 22 | DNAJC11 | 841 | 7.427 | 0.2446 | Yes | ||
| 23 | CDC45L | 960 | 6.993 | 0.2475 | Yes | ||
| 24 | ATF5 | 998 | 6.881 | 0.2540 | Yes | ||
| 25 | UNG | 1088 | 6.592 | 0.2578 | Yes | ||
| 26 | AP4M1 | 1145 | 6.445 | 0.2629 | Yes | ||
| 27 | E2F3 | 1330 | 5.964 | 0.2616 | Yes | ||
| 28 | E2F1 | 1414 | 5.788 | 0.2647 | Yes | ||
| 29 | POLA2 | 1425 | 5.762 | 0.2711 | Yes | ||
| 30 | CDCA7 | 1636 | 5.336 | 0.2678 | Yes | ||
| 31 | RAD51 | 1676 | 5.285 | 0.2724 | Yes | ||
| 32 | AK2 | 1717 | 5.217 | 0.2767 | Yes | ||
| 33 | KPNB1 | 1789 | 5.095 | 0.2796 | Yes | ||
| 34 | MCM3 | 1864 | 4.991 | 0.2821 | Yes | ||
| 35 | ILF3 | 1868 | 4.988 | 0.2880 | Yes | ||
| 36 | FANCD2 | 1920 | 4.913 | 0.2915 | Yes | ||
| 37 | CLSPN | 1930 | 4.901 | 0.2969 | Yes | ||
| 38 | CDC6 | 1934 | 4.895 | 0.3026 | Yes | ||
| 39 | EZH2 | 2009 | 4.793 | 0.3049 | Yes | ||
| 40 | UFD1L | 2122 | 4.650 | 0.3054 | Yes | ||
| 41 | PHF5A | 2139 | 4.624 | 0.3101 | Yes | ||
| 42 | PPP1R8 | 2224 | 4.511 | 0.3117 | Yes | ||
| 43 | CDC25A | 2359 | 4.348 | 0.3107 | Yes | ||
| 44 | PCNA | 2502 | 4.211 | 0.3092 | Yes | ||
| 45 | IPO7 | 2553 | 4.153 | 0.3119 | Yes | ||
| 46 | PKMYT1 | 2581 | 4.124 | 0.3156 | Yes | ||
| 47 | PAX6 | 2693 | 4.022 | 0.3153 | Yes | ||
| 48 | POLD1 | 2748 | 3.984 | 0.3176 | Yes | ||
| 49 | SLC38A1 | 2808 | 3.945 | 0.3196 | Yes | ||
| 50 | WEE1 | 2892 | 3.877 | 0.3204 | Yes | ||
| 51 | POLD3 | 2921 | 3.858 | 0.3237 | Yes | ||
| 52 | PVRL1 | 3000 | 3.798 | 0.3246 | Yes | ||
| 53 | NUP62 | 3002 | 3.797 | 0.3291 | Yes | ||
| 54 | CDT1 | 3024 | 3.784 | 0.3327 | Yes | ||
| 55 | HMGN2 | 3078 | 3.749 | 0.3347 | Yes | ||
| 56 | NELL2 | 3085 | 3.745 | 0.3389 | Yes | ||
| 57 | HS6ST3 | 3148 | 3.695 | 0.3405 | Yes | ||
| 58 | MCM2 | 3157 | 3.689 | 0.3445 | Yes | ||
| 59 | CTNND2 | 3222 | 3.638 | 0.3459 | Yes | ||
| 60 | POLE2 | 3339 | 3.542 | 0.3448 | Yes | ||
| 61 | SLC9A5 | 3367 | 3.520 | 0.3478 | Yes | ||
| 62 | DNMT1 | 3447 | 3.462 | 0.3483 | Yes | ||
| 63 | SFRS1 | 3608 | 3.355 | 0.3450 | No | ||
| 64 | TYRO3 | 3877 | 3.216 | 0.3365 | No | ||
| 65 | UXT | 3932 | 3.184 | 0.3378 | No | ||
| 66 | PHF15 | 4144 | 3.073 | 0.3318 | No | ||
| 67 | ATAD2 | 4214 | 3.035 | 0.3323 | No | ||
| 68 | PAQR4 | 4246 | 3.021 | 0.3345 | No | ||
| 69 | CSPG2 | 4395 | 2.954 | 0.3312 | No | ||
| 70 | ASXL2 | 4402 | 2.952 | 0.3344 | No | ||
| 71 | HTF9C | 4461 | 2.927 | 0.3353 | No | ||
| 72 | TMPO | 4551 | 2.891 | 0.3346 | No | ||
| 73 | NRP2 | 4700 | 2.836 | 0.3312 | No | ||
| 74 | MYH10 | 4767 | 2.808 | 0.3315 | No | ||
| 75 | KCND2 | 4780 | 2.799 | 0.3343 | No | ||
| 76 | SYNGR4 | 5243 | 2.641 | 0.3163 | No | ||
| 77 | PCSK1 | 5578 | 2.532 | 0.3040 | No | ||
| 78 | CBX5 | 5641 | 2.513 | 0.3041 | No | ||
| 79 | STMN1 | 5657 | 2.509 | 0.3064 | No | ||
| 80 | EFNA5 | 5947 | 2.415 | 0.2961 | No | ||
| 81 | RRM2 | 6132 | 2.358 | 0.2904 | No | ||
| 82 | MCM8 | 6691 | 2.199 | 0.2674 | No | ||
| 83 | MSH2 | 7101 | 2.090 | 0.2511 | No | ||
| 84 | ING3 | 7332 | 2.028 | 0.2430 | No | ||
| 85 | RPS6KA5 | 7337 | 2.027 | 0.2452 | No | ||
| 86 | ARHGAP6 | 7368 | 2.020 | 0.2463 | No | ||
| 87 | STAG2 | 7493 | 1.988 | 0.2430 | No | ||
| 88 | ADAMTS2 | 7554 | 1.973 | 0.2426 | No | ||
| 89 | GRIA4 | 7645 | 1.949 | 0.2408 | No | ||
| 90 | GABRB3 | 7813 | 1.903 | 0.2354 | No | ||
| 91 | SEMA5A | 8200 | 1.813 | 0.2198 | No | ||
| 92 | CORT | 8912 | 1.645 | 0.1891 | No | ||
| 93 | HIRA | 8916 | 1.644 | 0.1910 | No | ||
| 94 | ARHGAP11A | 8940 | 1.640 | 0.1919 | No | ||
| 95 | JPH1 | 8980 | 1.630 | 0.1920 | No | ||
| 96 | HNRPD | 9966 | 1.392 | 0.1485 | No | ||
| 97 | SLC6A4 | 10434 | 1.279 | 0.1285 | No | ||
| 98 | DMD | 10568 | 1.245 | 0.1239 | No | ||
| 99 | INSM1 | 12095 | 0.812 | 0.0548 | No | ||
| 100 | YBX2 | 12113 | 0.808 | 0.0550 | No | ||
| 101 | KCNA6 | 12307 | 0.751 | 0.0471 | No | ||
| 102 | EPHB1 | 12428 | 0.716 | 0.0424 | No | ||
| 103 | WNT3 | 12449 | 0.708 | 0.0423 | No | ||
| 104 | TREX2 | 12746 | 0.612 | 0.0295 | No | ||
| 105 | HOXC10 | 12756 | 0.605 | 0.0298 | No | ||
| 106 | FHOD1 | 12880 | 0.559 | 0.0248 | No | ||
| 107 | FBXO5 | 12981 | 0.520 | 0.0208 | No | ||
| 108 | RASAL2 | 13152 | 0.456 | 0.0136 | No | ||
| 109 | RHD | 13175 | 0.446 | 0.0131 | No | ||
| 110 | PODN | 13271 | 0.409 | 0.0092 | No | ||
| 111 | PIM1 | 13451 | 0.338 | 0.0014 | No | ||
| 112 | CASP8AP2 | 15356 | -0.775 | -0.0851 | No | ||
| 113 | MXD3 | 15419 | -0.824 | -0.0869 | No | ||
| 114 | EED | 15730 | -1.033 | -0.0999 | No | ||
| 115 | DDB2 | 15906 | -1.159 | -0.1066 | No | ||
| 116 | FMO4 | 16061 | -1.291 | -0.1121 | No | ||
| 117 | STK35 | 16140 | -1.357 | -0.1141 | No | ||
| 118 | PRKDC | 16339 | -1.517 | -0.1213 | No | ||
| 119 | BRMS1L | 16635 | -1.787 | -0.1328 | No | ||
| 120 | DNAJC5G | 16745 | -1.879 | -0.1355 | No | ||
| 121 | ABI2 | 16841 | -1.966 | -0.1375 | No | ||
| 122 | HCN3 | 16961 | -2.085 | -0.1405 | No | ||
| 123 | PHC1 | 17194 | -2.296 | -0.1484 | No | ||
| 124 | SFRS12 | 17306 | -2.399 | -0.1506 | No | ||
| 125 | GPRC5B | 17315 | -2.402 | -0.1481 | No | ||
| 126 | USP37 | 17763 | -2.848 | -0.1653 | No | ||
| 127 | SOAT1 | 17864 | -2.959 | -0.1663 | No | ||
| 128 | IL4I1 | 18167 | -3.291 | -0.1763 | No | ||
| 129 | ALDH6A1 | 18168 | -3.291 | -0.1723 | No | ||
| 130 | ARID4A | 18192 | -3.324 | -0.1694 | No | ||
| 131 | RAVER1 | 18505 | -3.665 | -0.1794 | No | ||
| 132 | POLE4 | 18516 | -3.673 | -0.1754 | No | ||
| 133 | HNRPUL1 | 18530 | -3.691 | -0.1716 | No | ||
| 134 | CCNT1 | 18554 | -3.710 | -0.1683 | No | ||
| 135 | PHF13 | 18563 | -3.718 | -0.1642 | No | ||
| 136 | MAZ | 18575 | -3.730 | -0.1602 | No | ||
| 137 | TBX6 | 18878 | -4.125 | -0.1692 | No | ||
| 138 | E2F7 | 19032 | -4.324 | -0.1710 | No | ||
| 139 | UGCGL1 | 19443 | -4.963 | -0.1839 | No | ||
| 140 | H2AFZ | 19510 | -5.048 | -0.1809 | No | ||
| 141 | ACO2 | 19662 | -5.276 | -0.1816 | No | ||
| 142 | HNRPR | 19749 | -5.403 | -0.1791 | No | ||
| 143 | ZCCHC8 | 19907 | -5.676 | -0.1795 | No | ||
| 144 | PPM1D | 20148 | -6.160 | -0.1832 | No | ||
| 145 | SUMO1 | 20226 | -6.338 | -0.1791 | No | ||
| 146 | DDX17 | 20343 | -6.577 | -0.1766 | No | ||
| 147 | ERBB2IP | 20406 | -6.743 | -0.1714 | No | ||
| 148 | SP3 | 20510 | -6.981 | -0.1678 | No | ||
| 149 | RAB11B | 20694 | -7.474 | -0.1673 | No | ||
| 150 | POLR2A | 20787 | -7.778 | -0.1622 | No | ||
| 151 | KBTBD7 | 20863 | -8.031 | -0.1560 | No | ||
| 152 | PPP1CC | 20962 | -8.421 | -0.1505 | No | ||
| 153 | ID3 | 21036 | -8.725 | -0.1434 | No | ||
| 154 | NUP153 | 21076 | -8.855 | -0.1346 | No | ||
| 155 | EPC1 | 21088 | -8.882 | -0.1245 | No | ||
| 156 | USP52 | 21092 | -8.896 | -0.1141 | No | ||
| 157 | SFRS10 | 21328 | -10.041 | -0.1129 | No | ||
| 158 | CHD2 | 21351 | -10.205 | -0.1017 | No | ||
| 159 | PB1 | 21513 | -11.385 | -0.0955 | No | ||
| 160 | GSPT1 | 21530 | -11.532 | -0.0825 | No | ||
| 161 | CTCF | 21662 | -13.154 | -0.0728 | No | ||
| 162 | SLCO3A1 | 21749 | -14.577 | -0.0593 | No | ||
| 163 | WHSC1L1 | 21772 | -15.077 | -0.0423 | No | ||
| 164 | ZBTB4 | 21848 | -17.473 | -0.0249 | No | ||
| 165 | DCK | 21929 | -24.615 | 0.0008 | No |