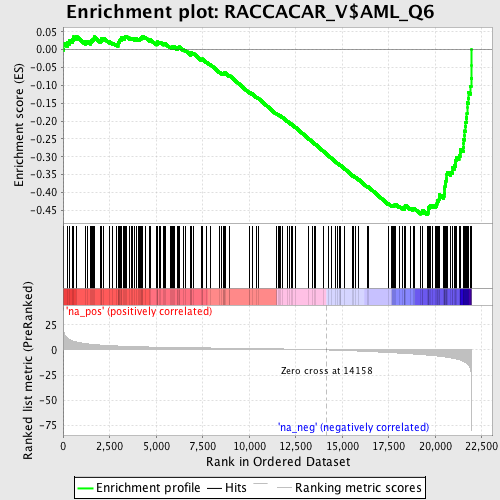
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RACCACAR_V$AML_Q6 |
| Enrichment Score (ES) | -0.46160027 |
| Normalized Enrichment Score (NES) | -1.9366094 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0061851195 |
| FWER p-Value | 0.029 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AMD1 | 32 | 18.200 | 0.0178 | No | ||
| 2 | EIF5A | 229 | 11.555 | 0.0211 | No | ||
| 3 | ARHGAP8 | 357 | 10.197 | 0.0261 | No | ||
| 4 | MSRA | 508 | 8.981 | 0.0287 | No | ||
| 5 | MTX1 | 561 | 8.626 | 0.0355 | No | ||
| 6 | ETF1 | 722 | 7.905 | 0.0365 | No | ||
| 7 | IL7R | 1187 | 6.326 | 0.0219 | No | ||
| 8 | MAFG | 1317 | 5.991 | 0.0223 | No | ||
| 9 | HEMGN | 1470 | 5.640 | 0.0213 | No | ||
| 10 | NUTF2 | 1511 | 5.580 | 0.0254 | No | ||
| 11 | FCER1G | 1598 | 5.415 | 0.0272 | No | ||
| 12 | RGMA | 1657 | 5.312 | 0.0302 | No | ||
| 13 | CCL2 | 1663 | 5.299 | 0.0356 | No | ||
| 14 | PPARG | 1999 | 4.807 | 0.0253 | No | ||
| 15 | ATP5A1 | 2058 | 4.738 | 0.0277 | No | ||
| 16 | LIF | 2069 | 4.729 | 0.0322 | No | ||
| 17 | RPP21 | 2191 | 4.550 | 0.0315 | No | ||
| 18 | OSCAR | 2481 | 4.228 | 0.0227 | No | ||
| 19 | PROK2 | 2669 | 4.044 | 0.0184 | No | ||
| 20 | PPP1R12C | 2848 | 3.909 | 0.0144 | No | ||
| 21 | HOXC6 | 2951 | 3.830 | 0.0137 | No | ||
| 22 | ZFHX1B | 2954 | 3.830 | 0.0177 | No | ||
| 23 | AGTRL1 | 2978 | 3.814 | 0.0207 | No | ||
| 24 | RIN1 | 3026 | 3.784 | 0.0226 | No | ||
| 25 | LUC7L | 3048 | 3.765 | 0.0256 | No | ||
| 26 | IL3 | 3065 | 3.756 | 0.0288 | No | ||
| 27 | RYBP | 3121 | 3.717 | 0.0303 | No | ||
| 28 | SDF2L1 | 3127 | 3.710 | 0.0340 | No | ||
| 29 | GABRA6 | 3221 | 3.639 | 0.0336 | No | ||
| 30 | TBX5 | 3322 | 3.557 | 0.0327 | No | ||
| 31 | PTPRS | 3327 | 3.551 | 0.0363 | No | ||
| 32 | CALR | 3379 | 3.515 | 0.0377 | No | ||
| 33 | ZIC4 | 3571 | 3.386 | 0.0325 | No | ||
| 34 | PCSK5 | 3657 | 3.329 | 0.0321 | No | ||
| 35 | SOX5 | 3742 | 3.284 | 0.0318 | No | ||
| 36 | HOXB6 | 3860 | 3.223 | 0.0298 | No | ||
| 37 | TULP4 | 3921 | 3.188 | 0.0304 | No | ||
| 38 | SUPT16H | 4044 | 3.117 | 0.0281 | No | ||
| 39 | NUCB2 | 4119 | 3.087 | 0.0280 | No | ||
| 40 | GTF2A1 | 4142 | 3.074 | 0.0303 | No | ||
| 41 | SREBF2 | 4187 | 3.050 | 0.0315 | No | ||
| 42 | EYA1 | 4201 | 3.043 | 0.0341 | No | ||
| 43 | AKAP3 | 4247 | 3.020 | 0.0353 | No | ||
| 44 | HLX1 | 4266 | 3.010 | 0.0376 | No | ||
| 45 | COMMD5 | 4428 | 2.940 | 0.0333 | No | ||
| 46 | SCRN1 | 4652 | 2.854 | 0.0261 | No | ||
| 47 | PTK9 | 4691 | 2.839 | 0.0274 | No | ||
| 48 | BMPR1B | 5009 | 2.718 | 0.0157 | No | ||
| 49 | CITED1 | 5063 | 2.698 | 0.0161 | No | ||
| 50 | ADAM28 | 5065 | 2.697 | 0.0189 | No | ||
| 51 | PDGFC | 5069 | 2.696 | 0.0217 | No | ||
| 52 | POU2F1 | 5182 | 2.660 | 0.0193 | No | ||
| 53 | GPR15 | 5236 | 2.643 | 0.0197 | No | ||
| 54 | OTP | 5388 | 2.596 | 0.0155 | No | ||
| 55 | TRPM8 | 5420 | 2.586 | 0.0169 | No | ||
| 56 | APOB48R | 5490 | 2.560 | 0.0164 | No | ||
| 57 | NAPSA | 5748 | 2.477 | 0.0072 | No | ||
| 58 | GPD1 | 5846 | 2.447 | 0.0054 | No | ||
| 59 | PPP1R14C | 5854 | 2.445 | 0.0076 | No | ||
| 60 | PGF | 5944 | 2.416 | 0.0061 | No | ||
| 61 | KRTAP6-3 | 5965 | 2.407 | 0.0077 | No | ||
| 62 | HOXB4 | 6138 | 2.354 | 0.0023 | No | ||
| 63 | BCAR3 | 6163 | 2.347 | 0.0037 | No | ||
| 64 | ATBF1 | 6181 | 2.341 | 0.0054 | No | ||
| 65 | PIK3R1 | 6230 | 2.327 | 0.0057 | No | ||
| 66 | IFNG | 6243 | 2.320 | 0.0076 | No | ||
| 67 | HHIP | 6461 | 2.262 | 0.0000 | No | ||
| 68 | CTHRC1 | 6570 | 2.232 | -0.0026 | No | ||
| 69 | FGFR1 | 6855 | 2.152 | -0.0133 | No | ||
| 70 | CHAD | 6864 | 2.150 | -0.0114 | No | ||
| 71 | TNFRSF12A | 6880 | 2.145 | -0.0098 | No | ||
| 72 | HOXC4 | 6893 | 2.143 | -0.0081 | No | ||
| 73 | ERG | 7011 | 2.116 | -0.0112 | No | ||
| 74 | C1QTNF6 | 7448 | 2.000 | -0.0292 | No | ||
| 75 | PDE6H | 7465 | 1.995 | -0.0278 | No | ||
| 76 | KCNJ1 | 7476 | 1.993 | -0.0261 | No | ||
| 77 | DAB2 | 7717 | 1.926 | -0.0351 | No | ||
| 78 | HOXA9 | 7895 | 1.884 | -0.0412 | No | ||
| 79 | HEY1 | 8405 | 1.769 | -0.0628 | No | ||
| 80 | SLC31A2 | 8534 | 1.735 | -0.0668 | No | ||
| 81 | ELAVL2 | 8601 | 1.718 | -0.0680 | No | ||
| 82 | THBS3 | 8639 | 1.707 | -0.0679 | No | ||
| 83 | LTBP1 | 8657 | 1.702 | -0.0669 | No | ||
| 84 | CCR1 | 8658 | 1.701 | -0.0651 | No | ||
| 85 | BNC2 | 8700 | 1.690 | -0.0652 | No | ||
| 86 | GPR155 | 8924 | 1.643 | -0.0737 | No | ||
| 87 | DMP1 | 8938 | 1.641 | -0.0725 | No | ||
| 88 | CPA2 | 10011 | 1.379 | -0.1203 | No | ||
| 89 | KCTD15 | 10018 | 1.378 | -0.1192 | No | ||
| 90 | SLC26A7 | 10152 | 1.346 | -0.1238 | No | ||
| 91 | HOXB3 | 10393 | 1.288 | -0.1335 | No | ||
| 92 | CCL4 | 10495 | 1.263 | -0.1368 | No | ||
| 93 | ADAMTS8 | 11444 | 1.009 | -0.1793 | No | ||
| 94 | WNT8B | 11451 | 1.007 | -0.1785 | No | ||
| 95 | NEK6 | 11571 | 0.974 | -0.1829 | No | ||
| 96 | ZBTB8 | 11637 | 0.955 | -0.1849 | No | ||
| 97 | SPIB | 11658 | 0.948 | -0.1848 | No | ||
| 98 | LIFR | 11793 | 0.908 | -0.1900 | No | ||
| 99 | GPR50 | 12066 | 0.822 | -0.2017 | No | ||
| 100 | DAB2IP | 12167 | 0.792 | -0.2054 | No | ||
| 101 | SLC36A2 | 12245 | 0.768 | -0.2081 | No | ||
| 102 | GPX1 | 12331 | 0.745 | -0.2113 | No | ||
| 103 | HOXB5 | 12488 | 0.696 | -0.2177 | No | ||
| 104 | POLG2 | 13208 | 0.431 | -0.2503 | No | ||
| 105 | NAV3 | 13380 | 0.365 | -0.2578 | No | ||
| 106 | MATN1 | 13529 | 0.304 | -0.2642 | No | ||
| 107 | CSF2 | 13564 | 0.288 | -0.2655 | No | ||
| 108 | RGS1 | 14005 | 0.079 | -0.2856 | No | ||
| 109 | OLIG3 | 14279 | -0.077 | -0.2981 | No | ||
| 110 | POF1B | 14401 | -0.155 | -0.3035 | No | ||
| 111 | CXCR3 | 14645 | -0.304 | -0.3143 | No | ||
| 112 | BATF | 14761 | -0.391 | -0.3192 | No | ||
| 113 | ENTPD3 | 14831 | -0.433 | -0.3219 | No | ||
| 114 | SCNN1A | 14906 | -0.481 | -0.3248 | No | ||
| 115 | REL | 14913 | -0.486 | -0.3246 | No | ||
| 116 | PAX1 | 15132 | -0.635 | -0.3339 | No | ||
| 117 | DKK1 | 15575 | -0.930 | -0.3532 | No | ||
| 118 | PPP2R3A | 15598 | -0.943 | -0.3533 | No | ||
| 119 | HOXD10 | 15719 | -1.027 | -0.3577 | No | ||
| 120 | PLXNC1 | 15885 | -1.144 | -0.3641 | No | ||
| 121 | RTN4RL1 | 16334 | -1.515 | -0.3830 | No | ||
| 122 | DYRK1B | 16393 | -1.569 | -0.3840 | No | ||
| 123 | FOXP3 | 17466 | -2.555 | -0.4306 | No | ||
| 124 | ITGB7 | 17644 | -2.731 | -0.4358 | No | ||
| 125 | RHOG | 17692 | -2.775 | -0.4351 | No | ||
| 126 | TACC2 | 17778 | -2.868 | -0.4359 | No | ||
| 127 | ASB7 | 17825 | -2.925 | -0.4349 | No | ||
| 128 | PTPN22 | 17874 | -2.968 | -0.4340 | No | ||
| 129 | SIRT1 | 18076 | -3.186 | -0.4398 | No | ||
| 130 | RBBP6 | 18221 | -3.350 | -0.4429 | No | ||
| 131 | MAP1LC3A | 18329 | -3.453 | -0.4442 | No | ||
| 132 | CD6 | 18354 | -3.478 | -0.4416 | No | ||
| 133 | ACIN1 | 18367 | -3.498 | -0.4384 | No | ||
| 134 | BLOC1S1 | 18395 | -3.528 | -0.4359 | No | ||
| 135 | RPA3 | 18674 | -3.847 | -0.4446 | No | ||
| 136 | SFRS4 | 18800 | -3.999 | -0.4461 | No | ||
| 137 | ABHD8 | 18856 | -4.087 | -0.4443 | No | ||
| 138 | WAS | 19208 | -4.589 | -0.4555 | No | ||
| 139 | MTMR4 | 19309 | -4.743 | -0.4551 | No | ||
| 140 | WBP1 | 19312 | -4.751 | -0.4502 | No | ||
| 141 | DGKZ | 19562 | -5.140 | -0.4561 | Yes | ||
| 142 | EIF2C1 | 19610 | -5.207 | -0.4528 | Yes | ||
| 143 | EVI2A | 19614 | -5.215 | -0.4474 | Yes | ||
| 144 | TRPV2 | 19638 | -5.253 | -0.4429 | Yes | ||
| 145 | NDUFA3 | 19698 | -5.335 | -0.4399 | Yes | ||
| 146 | RUNX2 | 19721 | -5.364 | -0.4352 | Yes | ||
| 147 | ARHGEF2 | 19872 | -5.614 | -0.4362 | Yes | ||
| 148 | MKLN1 | 19997 | -5.866 | -0.4356 | Yes | ||
| 149 | UBE2R2 | 20060 | -6.001 | -0.4321 | Yes | ||
| 150 | TBL1X | 20091 | -6.058 | -0.4271 | Yes | ||
| 151 | RUNX1 | 20112 | -6.091 | -0.4215 | Yes | ||
| 152 | EMP1 | 20196 | -6.261 | -0.4187 | Yes | ||
| 153 | AKT2 | 20209 | -6.297 | -0.4126 | Yes | ||
| 154 | CDC37L1 | 20219 | -6.321 | -0.4063 | Yes | ||
| 155 | CHES1 | 20432 | -6.813 | -0.4088 | Yes | ||
| 156 | TBPL1 | 20470 | -6.907 | -0.4031 | Yes | ||
| 157 | MSI2 | 20493 | -6.952 | -0.3968 | Yes | ||
| 158 | AGPAT4 | 20495 | -6.955 | -0.3894 | Yes | ||
| 159 | CCNT2 | 20516 | -6.990 | -0.3829 | Yes | ||
| 160 | EDARADD | 20526 | -7.013 | -0.3759 | Yes | ||
| 161 | SOST | 20557 | -7.092 | -0.3698 | Yes | ||
| 162 | TCF7 | 20577 | -7.144 | -0.3630 | Yes | ||
| 163 | SLA2 | 20618 | -7.270 | -0.3572 | Yes | ||
| 164 | PHF6 | 20621 | -7.281 | -0.3495 | Yes | ||
| 165 | AP1S2 | 20651 | -7.362 | -0.3430 | Yes | ||
| 166 | HPCAL1 | 20837 | -7.938 | -0.3431 | Yes | ||
| 167 | CPNE1 | 20927 | -8.271 | -0.3384 | Yes | ||
| 168 | PITPNC1 | 20931 | -8.288 | -0.3298 | Yes | ||
| 169 | ID3 | 21036 | -8.725 | -0.3253 | Yes | ||
| 170 | FLI1 | 21086 | -8.875 | -0.3181 | Yes | ||
| 171 | JMJD1C | 21097 | -8.917 | -0.3091 | Yes | ||
| 172 | ATF7IP | 21131 | -9.063 | -0.3010 | Yes | ||
| 173 | MADD | 21274 | -9.747 | -0.2972 | Yes | ||
| 174 | PICALM | 21330 | -10.065 | -0.2890 | Yes | ||
| 175 | IL17RE | 21341 | -10.123 | -0.2788 | Yes | ||
| 176 | MTMR3 | 21499 | -11.309 | -0.2740 | Yes | ||
| 177 | TAGAP | 21515 | -11.396 | -0.2626 | Yes | ||
| 178 | RASSF2 | 21521 | -11.453 | -0.2506 | Yes | ||
| 179 | DOK2 | 21579 | -12.057 | -0.2405 | Yes | ||
| 180 | PAFAH1B1 | 21585 | -12.135 | -0.2278 | Yes | ||
| 181 | CD69 | 21606 | -12.413 | -0.2155 | Yes | ||
| 182 | TCF12 | 21627 | -12.637 | -0.2031 | Yes | ||
| 183 | RGS14 | 21660 | -13.135 | -0.1906 | Yes | ||
| 184 | PSCDBP | 21699 | -13.765 | -0.1777 | Yes | ||
| 185 | CBL | 21707 | -13.898 | -0.1633 | Yes | ||
| 186 | SLC12A6 | 21710 | -13.946 | -0.1486 | Yes | ||
| 187 | LCK | 21778 | -15.166 | -0.1356 | Yes | ||
| 188 | TLN1 | 21795 | -15.630 | -0.1197 | Yes | ||
| 189 | CD28 | 21902 | -20.380 | -0.1029 | Yes | ||
| 190 | CRY1 | 21920 | -22.141 | -0.0802 | Yes | ||
| 191 | SCN2B | 21941 | -34.007 | -0.0450 | Yes | ||
| 192 | BCL6 | 21944 | -42.648 | 0.0001 | Yes |