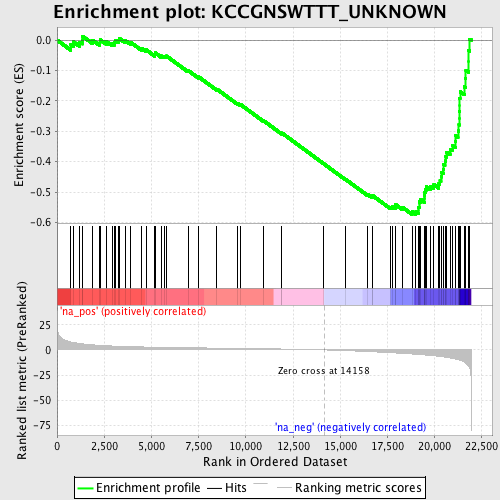
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | KCCGNSWTTT_UNKNOWN |
| Enrichment Score (ES) | -0.5739562 |
| Normalized Enrichment Score (NES) | -2.1443763 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0025529817 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CRK | 726 | 7.882 | -0.0157 | No | ||
| 2 | SYNCRIP | 870 | 7.330 | -0.0059 | No | ||
| 3 | HNRPK | 1201 | 6.287 | -0.0070 | No | ||
| 4 | E2F3 | 1330 | 5.964 | 0.0005 | No | ||
| 5 | XPO1 | 1342 | 5.926 | 0.0131 | No | ||
| 6 | ILF3 | 1868 | 4.988 | 0.0002 | No | ||
| 7 | MAX | 2271 | 4.449 | -0.0082 | No | ||
| 8 | EIF4G2 | 2275 | 4.447 | 0.0015 | No | ||
| 9 | BCL11A | 2627 | 4.079 | -0.0054 | No | ||
| 10 | SFRS3 | 2925 | 3.854 | -0.0104 | No | ||
| 11 | DHX15 | 3055 | 3.762 | -0.0080 | No | ||
| 12 | LPHN3 | 3067 | 3.754 | -0.0001 | No | ||
| 13 | HNRPA2B1 | 3240 | 3.624 | 0.0001 | No | ||
| 14 | CCNA2 | 3305 | 3.575 | 0.0051 | No | ||
| 15 | ATP5G2 | 3603 | 3.359 | -0.0010 | No | ||
| 16 | PTMA | 3909 | 3.196 | -0.0078 | No | ||
| 17 | GNG11 | 4495 | 2.916 | -0.0281 | No | ||
| 18 | TDRD3 | 4711 | 2.830 | -0.0316 | No | ||
| 19 | POU2F1 | 5182 | 2.660 | -0.0472 | No | ||
| 20 | PRICKLE1 | 5195 | 2.655 | -0.0418 | No | ||
| 21 | ASCL1 | 5545 | 2.543 | -0.0521 | No | ||
| 22 | PPP2R2B | 5684 | 2.500 | -0.0528 | No | ||
| 23 | SOS1 | 5789 | 2.466 | -0.0521 | No | ||
| 24 | IRX5 | 6943 | 2.132 | -0.1001 | No | ||
| 25 | STAG2 | 7493 | 1.988 | -0.1208 | No | ||
| 26 | PAK7 | 8468 | 1.750 | -0.1614 | No | ||
| 27 | CBX3 | 9582 | 1.486 | -0.2090 | No | ||
| 28 | AKAP8 | 9724 | 1.453 | -0.2122 | No | ||
| 29 | NHS | 10933 | 1.152 | -0.2649 | No | ||
| 30 | FOXA2 | 11905 | 0.869 | -0.3074 | No | ||
| 31 | OTX2 | 11908 | 0.868 | -0.3055 | No | ||
| 32 | DAP3 | 14129 | 0.012 | -0.4070 | No | ||
| 33 | CXXC5 | 15268 | -0.719 | -0.4575 | No | ||
| 34 | HNRPH1 | 16425 | -1.592 | -0.5068 | No | ||
| 35 | ACLY | 16699 | -1.839 | -0.5152 | No | ||
| 36 | PTBP2 | 16729 | -1.863 | -0.5124 | No | ||
| 37 | LRRC1 | 17673 | -2.753 | -0.5494 | No | ||
| 38 | TAF15 | 17762 | -2.847 | -0.5470 | No | ||
| 39 | NCAM1 | 17916 | -3.014 | -0.5473 | No | ||
| 40 | AP1G1 | 17936 | -3.039 | -0.5414 | No | ||
| 41 | ADNP | 18312 | -3.434 | -0.5509 | No | ||
| 42 | EIF4A2 | 18810 | -4.014 | -0.5647 | Yes | ||
| 43 | DDX6 | 19013 | -4.300 | -0.5644 | Yes | ||
| 44 | SF1 | 19149 | -4.500 | -0.5605 | Yes | ||
| 45 | UBC | 19151 | -4.504 | -0.5506 | Yes | ||
| 46 | PCF11 | 19184 | -4.553 | -0.5419 | Yes | ||
| 47 | JMJD1A | 19185 | -4.554 | -0.5317 | Yes | ||
| 48 | ZFP91 | 19257 | -4.657 | -0.5246 | Yes | ||
| 49 | DDX5 | 19454 | -4.978 | -0.5225 | Yes | ||
| 50 | FOXO3A | 19458 | -4.980 | -0.5115 | Yes | ||
| 51 | LNPEP | 19484 | -5.011 | -0.5015 | Yes | ||
| 52 | FBXL11 | 19518 | -5.063 | -0.4918 | Yes | ||
| 53 | CSNK1A1 | 19558 | -5.138 | -0.4821 | Yes | ||
| 54 | FAAH | 19799 | -5.490 | -0.4809 | Yes | ||
| 55 | RFX1 | 19965 | -5.815 | -0.4755 | Yes | ||
| 56 | AP3D1 | 20227 | -6.341 | -0.4733 | Yes | ||
| 57 | WDR33 | 20288 | -6.468 | -0.4616 | Yes | ||
| 58 | DDX17 | 20343 | -6.577 | -0.4495 | Yes | ||
| 59 | ENSA | 20371 | -6.643 | -0.4359 | Yes | ||
| 60 | USP3 | 20458 | -6.887 | -0.4245 | Yes | ||
| 61 | OGT | 20476 | -6.916 | -0.4099 | Yes | ||
| 62 | CRKL | 20555 | -7.087 | -0.3977 | Yes | ||
| 63 | SPRED2 | 20599 | -7.209 | -0.3836 | Yes | ||
| 64 | SYT11 | 20635 | -7.316 | -0.3689 | Yes | ||
| 65 | RBPSUH | 20825 | -7.896 | -0.3600 | Yes | ||
| 66 | RBM4 | 20935 | -8.316 | -0.3464 | Yes | ||
| 67 | NONO | 21115 | -8.993 | -0.3346 | Yes | ||
| 68 | SRRM1 | 21127 | -9.050 | -0.3149 | Yes | ||
| 69 | DMTF1 | 21245 | -9.609 | -0.2989 | Yes | ||
| 70 | CUGBP1 | 21293 | -9.860 | -0.2791 | Yes | ||
| 71 | CNOT3 | 21304 | -9.928 | -0.2574 | Yes | ||
| 72 | ARMCX2 | 21308 | -9.942 | -0.2354 | Yes | ||
| 73 | SFRS10 | 21328 | -10.041 | -0.2140 | Yes | ||
| 74 | PICALM | 21330 | -10.065 | -0.1916 | Yes | ||
| 75 | RGL2 | 21354 | -10.219 | -0.1699 | Yes | ||
| 76 | PAFAH1B1 | 21585 | -12.135 | -0.1534 | Yes | ||
| 77 | MLL5 | 21645 | -12.951 | -0.1273 | Yes | ||
| 78 | CTCF | 21662 | -13.154 | -0.0987 | Yes | ||
| 79 | SP1 | 21811 | -15.984 | -0.0699 | Yes | ||
| 80 | YWHAE | 21814 | -16.134 | -0.0340 | Yes | ||
| 81 | JARID2 | 21867 | -18.002 | 0.0037 | Yes |