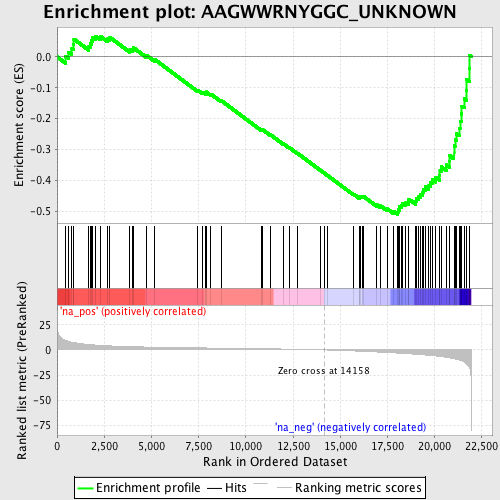
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | AAGWWRNYGGC_UNKNOWN |
| Enrichment Score (ES) | -0.5108664 |
| Normalized Enrichment Score (NES) | -1.9057993 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004729066 |
| FWER p-Value | 0.037 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARL1 | 451 | 9.321 | 0.0023 | No | ||
| 2 | NASP | 620 | 8.327 | 0.0151 | No | ||
| 3 | RNF26 | 782 | 7.655 | 0.0266 | No | ||
| 4 | RFC1 | 874 | 7.311 | 0.0404 | No | ||
| 5 | MRPL38 | 891 | 7.249 | 0.0575 | No | ||
| 6 | SMYD5 | 1685 | 5.270 | 0.0342 | No | ||
| 7 | CDKN2C | 1747 | 5.171 | 0.0442 | No | ||
| 8 | RNF12 | 1828 | 5.042 | 0.0529 | No | ||
| 9 | ATF1 | 1894 | 4.952 | 0.0621 | No | ||
| 10 | UBE2D3 | 2039 | 4.762 | 0.0673 | No | ||
| 11 | PSMD8 | 2299 | 4.418 | 0.0663 | No | ||
| 12 | PAX6 | 2693 | 4.022 | 0.0582 | No | ||
| 13 | RHOB | 2798 | 3.952 | 0.0632 | No | ||
| 14 | HOXB6 | 3860 | 3.223 | 0.0226 | No | ||
| 15 | APBA1 | 4006 | 3.142 | 0.0237 | No | ||
| 16 | SUPT16H | 4044 | 3.117 | 0.0297 | No | ||
| 17 | SUV39H2 | 4717 | 2.828 | 0.0059 | No | ||
| 18 | CRSP8 | 5152 | 2.669 | -0.0074 | No | ||
| 19 | RAD23A | 7460 | 1.997 | -0.1080 | No | ||
| 20 | SLITRK3 | 7711 | 1.928 | -0.1147 | No | ||
| 21 | IRAK1 | 7859 | 1.892 | -0.1167 | No | ||
| 22 | HOXA9 | 7895 | 1.884 | -0.1137 | No | ||
| 23 | PIGN | 8140 | 1.829 | -0.1203 | No | ||
| 24 | BNC2 | 8700 | 1.690 | -0.1418 | No | ||
| 25 | ADAMTS9 | 10806 | 1.186 | -0.2351 | No | ||
| 26 | CLSTN1 | 10864 | 1.170 | -0.2348 | No | ||
| 27 | RIF1 | 11316 | 1.050 | -0.2529 | No | ||
| 28 | LRRN3 | 11997 | 0.843 | -0.2819 | No | ||
| 29 | NEUROD1 | 12288 | 0.757 | -0.2933 | No | ||
| 30 | HOXC10 | 12756 | 0.605 | -0.3132 | No | ||
| 31 | NKIRAS2 | 13932 | 0.116 | -0.3666 | No | ||
| 32 | SFN | 14174 | -0.015 | -0.3776 | No | ||
| 33 | OTX1 | 14353 | -0.128 | -0.3854 | No | ||
| 34 | HOXA2 | 15700 | -1.015 | -0.4445 | No | ||
| 35 | CDC42BPB | 16031 | -1.265 | -0.4565 | No | ||
| 36 | POLK | 16048 | -1.280 | -0.4540 | No | ||
| 37 | PPM1E | 16058 | -1.289 | -0.4513 | No | ||
| 38 | NFYC | 16190 | -1.402 | -0.4538 | No | ||
| 39 | CXXC1 | 16245 | -1.443 | -0.4527 | No | ||
| 40 | PRDM1 | 16946 | -2.070 | -0.4797 | No | ||
| 41 | FMO5 | 17121 | -2.238 | -0.4821 | No | ||
| 42 | RALA | 17496 | -2.599 | -0.4928 | No | ||
| 43 | RAB1B | 17847 | -2.944 | -0.5016 | No | ||
| 44 | TIA1 | 18051 | -3.154 | -0.5031 | Yes | ||
| 45 | RBM3 | 18069 | -3.176 | -0.4961 | Yes | ||
| 46 | ARF1 | 18135 | -3.249 | -0.4910 | Yes | ||
| 47 | RFX3 | 18149 | -3.273 | -0.4836 | Yes | ||
| 48 | ANKRD28 | 18262 | -3.392 | -0.4804 | Yes | ||
| 49 | EIF5 | 18319 | -3.440 | -0.4745 | Yes | ||
| 50 | STRN4 | 18485 | -3.641 | -0.4730 | Yes | ||
| 51 | BCL2L2 | 18632 | -3.791 | -0.4704 | Yes | ||
| 52 | PPP1R12B | 18636 | -3.794 | -0.4612 | Yes | ||
| 53 | SNX13 | 19009 | -4.292 | -0.4676 | Yes | ||
| 54 | ANKHD1 | 19026 | -4.321 | -0.4577 | Yes | ||
| 55 | RPS27 | 19141 | -4.489 | -0.4519 | Yes | ||
| 56 | ARCN1 | 19240 | -4.636 | -0.4450 | Yes | ||
| 57 | PLAG1 | 19351 | -4.812 | -0.4382 | Yes | ||
| 58 | DNAJC7 | 19402 | -4.899 | -0.4284 | Yes | ||
| 59 | FKRP | 19501 | -5.036 | -0.4205 | Yes | ||
| 60 | CHCHD7 | 19703 | -5.341 | -0.4165 | Yes | ||
| 61 | CHD4 | 19790 | -5.476 | -0.4070 | Yes | ||
| 62 | COL4A3BP | 19878 | -5.631 | -0.3971 | Yes | ||
| 63 | UBE3A | 20067 | -6.019 | -0.3909 | Yes | ||
| 64 | VAMP2 | 20274 | -6.433 | -0.3845 | Yes | ||
| 65 | WDR33 | 20288 | -6.468 | -0.3692 | Yes | ||
| 66 | CNOT4 | 20347 | -6.588 | -0.3556 | Yes | ||
| 67 | TRIM8 | 20623 | -7.283 | -0.3503 | Yes | ||
| 68 | EP300 | 20781 | -7.751 | -0.3384 | Yes | ||
| 69 | POLR2A | 20787 | -7.778 | -0.3195 | Yes | ||
| 70 | PPARBP | 21031 | -8.705 | -0.3091 | Yes | ||
| 71 | GNAI2 | 21032 | -8.712 | -0.2877 | Yes | ||
| 72 | EPC1 | 21088 | -8.882 | -0.2684 | Yes | ||
| 73 | NRAS | 21157 | -9.167 | -0.2489 | Yes | ||
| 74 | CNOT3 | 21304 | -9.928 | -0.2312 | Yes | ||
| 75 | TSC1 | 21353 | -10.209 | -0.2082 | Yes | ||
| 76 | ZZEF1 | 21415 | -10.644 | -0.1848 | Yes | ||
| 77 | SFPQ | 21450 | -10.874 | -0.1596 | Yes | ||
| 78 | RAB22A | 21572 | -11.959 | -0.1357 | Yes | ||
| 79 | PHF8 | 21692 | -13.659 | -0.1076 | Yes | ||
| 80 | YY1 | 21700 | -13.773 | -0.0740 | Yes | ||
| 81 | MYLIP | 21841 | -17.145 | -0.0382 | Yes | ||
| 82 | ZBTB4 | 21848 | -17.473 | 0.0045 | Yes |