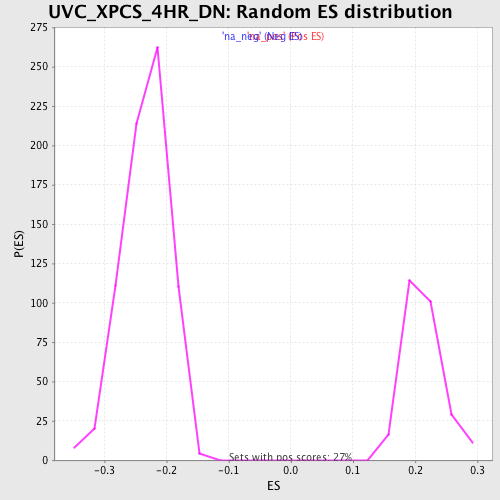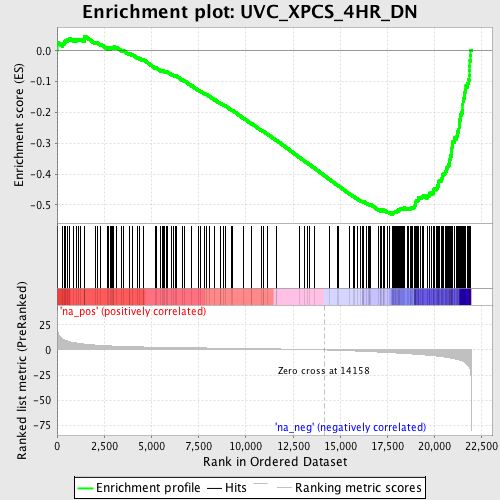
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | UVC_XPCS_4HR_DN |
| Enrichment Score (ES) | -0.53102165 |
| Normalized Enrichment Score (NES) | -2.2802155 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.2921392E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PBX3 | 2 | 31.298 | 0.0280 | No | ||
| 2 | WDR43 | 309 | 10.676 | 0.0235 | No | ||
| 3 | CYR61 | 367 | 10.135 | 0.0300 | No | ||
| 4 | MTAP | 447 | 9.352 | 0.0348 | No | ||
| 5 | SOCS5 | 568 | 8.590 | 0.0370 | No | ||
| 6 | TOPBP1 | 674 | 8.102 | 0.0395 | No | ||
| 7 | MYC | 869 | 7.330 | 0.0371 | No | ||
| 8 | BLM | 1034 | 6.781 | 0.0357 | No | ||
| 9 | TRIM2 | 1111 | 6.518 | 0.0380 | No | ||
| 10 | SHB | 1242 | 6.175 | 0.0376 | No | ||
| 11 | FGF5 | 1436 | 5.737 | 0.0339 | No | ||
| 12 | FBXL7 | 1439 | 5.723 | 0.0389 | No | ||
| 13 | EGFR | 1441 | 5.720 | 0.0440 | No | ||
| 14 | BAZ1A | 1472 | 5.636 | 0.0477 | No | ||
| 15 | CENTG2 | 2017 | 4.785 | 0.0270 | No | ||
| 16 | TRIO | 2114 | 4.664 | 0.0267 | No | ||
| 17 | PTPRK | 2304 | 4.414 | 0.0220 | No | ||
| 18 | SWAP70 | 2690 | 4.026 | 0.0079 | No | ||
| 19 | EPHA4 | 2706 | 4.014 | 0.0108 | No | ||
| 20 | CTBP2 | 2839 | 3.915 | 0.0083 | No | ||
| 21 | KNTC2 | 2867 | 3.894 | 0.0105 | No | ||
| 22 | ZFHX1B | 2954 | 3.830 | 0.0100 | No | ||
| 23 | ACVR1 | 3012 | 3.791 | 0.0108 | No | ||
| 24 | SAMD4A | 3013 | 3.791 | 0.0142 | No | ||
| 25 | PLEKHC1 | 3143 | 3.699 | 0.0116 | No | ||
| 26 | CENPE | 3418 | 3.481 | 0.0021 | No | ||
| 27 | DDX10 | 3519 | 3.426 | 0.0006 | No | ||
| 28 | CDK8 | 3816 | 3.244 | -0.0101 | No | ||
| 29 | HOXB2 | 3836 | 3.237 | -0.0081 | No | ||
| 30 | RRAS2 | 3969 | 3.162 | -0.0113 | No | ||
| 31 | LHFPL2 | 4284 | 3.002 | -0.0230 | No | ||
| 32 | CTGF | 4384 | 2.960 | -0.0249 | No | ||
| 33 | IRAK1BP1 | 4553 | 2.891 | -0.0301 | No | ||
| 34 | BMPR1A | 4580 | 2.883 | -0.0287 | No | ||
| 35 | PRR3 | 5228 | 2.644 | -0.0561 | No | ||
| 36 | RSN | 5263 | 2.635 | -0.0553 | No | ||
| 37 | WWC1 | 5498 | 2.557 | -0.0637 | No | ||
| 38 | ARHGAP29 | 5598 | 2.526 | -0.0660 | No | ||
| 39 | STK3 | 5627 | 2.519 | -0.0650 | No | ||
| 40 | DOCK4 | 5715 | 2.491 | -0.0668 | No | ||
| 41 | SOS1 | 5789 | 2.466 | -0.0679 | No | ||
| 42 | DLC1 | 5832 | 2.454 | -0.0677 | No | ||
| 43 | CEP135 | 6071 | 2.374 | -0.0765 | No | ||
| 44 | BCAR3 | 6163 | 2.347 | -0.0786 | No | ||
| 45 | VEGFC | 6286 | 2.307 | -0.0821 | No | ||
| 46 | SATB2 | 6312 | 2.302 | -0.0812 | No | ||
| 47 | WNT5A | 6638 | 2.213 | -0.0941 | No | ||
| 48 | FGF2 | 6738 | 2.186 | -0.0967 | No | ||
| 49 | ARFGEF1 | 7130 | 2.083 | -0.1128 | No | ||
| 50 | NFIB | 7485 | 1.991 | -0.1273 | No | ||
| 51 | DDEF2 | 7600 | 1.964 | -0.1308 | No | ||
| 52 | ORC5L | 7806 | 1.904 | -0.1385 | No | ||
| 53 | ACSL3 | 7897 | 1.883 | -0.1410 | No | ||
| 54 | SRGAP2 | 8054 | 1.846 | -0.1465 | No | ||
| 55 | PVRL3 | 8357 | 1.778 | -0.1588 | No | ||
| 56 | IL1RAP | 8649 | 1.705 | -0.1707 | No | ||
| 57 | MID1 | 8664 | 1.700 | -0.1698 | No | ||
| 58 | TGFB2 | 8826 | 1.665 | -0.1757 | No | ||
| 59 | HIRA | 8916 | 1.644 | -0.1783 | No | ||
| 60 | SCHIP1 | 9249 | 1.563 | -0.1922 | No | ||
| 61 | BICD1 | 9308 | 1.551 | -0.1934 | No | ||
| 62 | GLRB | 9900 | 1.410 | -0.2194 | No | ||
| 63 | GNAI1 | 10304 | 1.311 | -0.2367 | No | ||
| 64 | CENPC1 | 10306 | 1.310 | -0.2356 | No | ||
| 65 | ZZZ3 | 10839 | 1.178 | -0.2590 | No | ||
| 66 | PHLPPL | 10927 | 1.154 | -0.2620 | No | ||
| 67 | MTF2 | 10958 | 1.146 | -0.2623 | No | ||
| 68 | STARD13 | 11150 | 1.096 | -0.2701 | No | ||
| 69 | ZFYVE9 | 11608 | 0.964 | -0.2903 | No | ||
| 70 | BARD1 | 11636 | 0.955 | -0.2907 | No | ||
| 71 | NUAK1 | 12866 | 0.562 | -0.3467 | No | ||
| 72 | PARG | 13122 | 0.468 | -0.3580 | No | ||
| 73 | BHLHB2 | 13284 | 0.403 | -0.3651 | No | ||
| 74 | NAV3 | 13380 | 0.365 | -0.3691 | No | ||
| 75 | IL6 | 13644 | 0.251 | -0.3810 | No | ||
| 76 | KIAA1128 | 14455 | -0.187 | -0.4181 | No | ||
| 77 | F3 | 14869 | -0.457 | -0.4367 | No | ||
| 78 | LARP5 | 14927 | -0.492 | -0.4388 | No | ||
| 79 | UPF2 | 15475 | -0.862 | -0.4632 | No | ||
| 80 | RGS7 | 15691 | -1.007 | -0.4722 | No | ||
| 81 | EIF2C2 | 15736 | -1.037 | -0.4733 | No | ||
| 82 | RNGTT | 15917 | -1.169 | -0.4805 | No | ||
| 83 | KLF10 | 16085 | -1.313 | -0.4870 | No | ||
| 84 | ARHGAP12 | 16162 | -1.379 | -0.4893 | No | ||
| 85 | APRIN | 16177 | -1.392 | -0.4887 | No | ||
| 86 | CD2AP | 16239 | -1.438 | -0.4902 | No | ||
| 87 | DAPK1 | 16250 | -1.448 | -0.4894 | No | ||
| 88 | RAPGEF2 | 16380 | -1.556 | -0.4939 | No | ||
| 89 | EHBP1 | 16501 | -1.660 | -0.4979 | No | ||
| 90 | E2F5 | 16561 | -1.721 | -0.4991 | No | ||
| 91 | CAMSAP1L1 | 16565 | -1.723 | -0.4977 | No | ||
| 92 | SOS2 | 16589 | -1.746 | -0.4972 | No | ||
| 93 | TLE1 | 17016 | -2.145 | -0.5148 | No | ||
| 94 | NCK1 | 17148 | -2.262 | -0.5188 | No | ||
| 95 | CDYL | 17157 | -2.270 | -0.5172 | No | ||
| 96 | PHLPP | 17173 | -2.284 | -0.5158 | No | ||
| 97 | TIAM1 | 17271 | -2.359 | -0.5182 | No | ||
| 98 | ASXL1 | 17275 | -2.364 | -0.5162 | No | ||
| 99 | FCHSD2 | 17338 | -2.428 | -0.5168 | No | ||
| 100 | AKAP9 | 17533 | -2.631 | -0.5234 | No | ||
| 101 | CENPA | 17639 | -2.727 | -0.5258 | No | ||
| 102 | BTRC | 17754 | -2.835 | -0.5285 | Yes | ||
| 103 | TERF1 | 17758 | -2.839 | -0.5261 | Yes | ||
| 104 | CEP170 | 17803 | -2.895 | -0.5255 | Yes | ||
| 105 | CCDC93 | 17812 | -2.906 | -0.5232 | Yes | ||
| 106 | ANKS1A | 17870 | -2.963 | -0.5232 | Yes | ||
| 107 | REV3L | 17923 | -3.021 | -0.5229 | Yes | ||
| 108 | PIP5K3 | 17927 | -3.024 | -0.5203 | Yes | ||
| 109 | CLASP2 | 17995 | -3.100 | -0.5206 | Yes | ||
| 110 | TCF7L2 | 18032 | -3.133 | -0.5194 | Yes | ||
| 111 | TUBGCP3 | 18070 | -3.176 | -0.5183 | Yes | ||
| 112 | KIF2A | 18083 | -3.195 | -0.5160 | Yes | ||
| 113 | FRYL | 18121 | -3.241 | -0.5148 | Yes | ||
| 114 | ATP2B1 | 18161 | -3.285 | -0.5136 | Yes | ||
| 115 | ZHX3 | 18212 | -3.340 | -0.5129 | Yes | ||
| 116 | ANKRD17 | 18261 | -3.392 | -0.5121 | Yes | ||
| 117 | ZNF148 | 18296 | -3.423 | -0.5105 | Yes | ||
| 118 | FOXK2 | 18381 | -3.517 | -0.5112 | Yes | ||
| 119 | THRAP1 | 18415 | -3.552 | -0.5096 | Yes | ||
| 120 | EDD1 | 18562 | -3.718 | -0.5130 | Yes | ||
| 121 | UBXD7 | 18625 | -3.782 | -0.5124 | Yes | ||
| 122 | ZHX2 | 18710 | -3.894 | -0.5128 | Yes | ||
| 123 | FOXJ3 | 18730 | -3.918 | -0.5101 | Yes | ||
| 124 | FNDC3A | 18769 | -3.963 | -0.5083 | Yes | ||
| 125 | CHSY1 | 18818 | -4.020 | -0.5069 | Yes | ||
| 126 | CEP350 | 18911 | -4.164 | -0.5074 | Yes | ||
| 127 | STAM | 18942 | -4.204 | -0.5050 | Yes | ||
| 128 | CREB1 | 18962 | -4.227 | -0.5021 | Yes | ||
| 129 | TSPAN5 | 18976 | -4.237 | -0.4989 | Yes | ||
| 130 | SLC25A12 | 19006 | -4.291 | -0.4963 | Yes | ||
| 131 | SNX13 | 19009 | -4.292 | -0.4926 | Yes | ||
| 132 | CNOT2 | 19015 | -4.303 | -0.4889 | Yes | ||
| 133 | PTPN12 | 19048 | -4.339 | -0.4865 | Yes | ||
| 134 | KLF6 | 19095 | -4.409 | -0.4847 | Yes | ||
| 135 | IFRD1 | 19138 | -4.480 | -0.4826 | Yes | ||
| 136 | MARK3 | 19140 | -4.484 | -0.4786 | Yes | ||
| 137 | RCOR1 | 19148 | -4.497 | -0.4749 | Yes | ||
| 138 | WAPAL | 19275 | -4.690 | -0.4765 | Yes | ||
| 139 | EXT1 | 19335 | -4.786 | -0.4749 | Yes | ||
| 140 | CENTB2 | 19378 | -4.857 | -0.4724 | Yes | ||
| 141 | PUM2 | 19426 | -4.934 | -0.4702 | Yes | ||
| 142 | CBLB | 19603 | -5.196 | -0.4736 | Yes | ||
| 143 | TAF4 | 19624 | -5.239 | -0.4698 | Yes | ||
| 144 | ATF2 | 19713 | -5.352 | -0.4690 | Yes | ||
| 145 | USP15 | 19714 | -5.353 | -0.4642 | Yes | ||
| 146 | NRIP1 | 19757 | -5.420 | -0.4613 | Yes | ||
| 147 | ZNF292 | 19847 | -5.566 | -0.4604 | Yes | ||
| 148 | ITSN2 | 19939 | -5.749 | -0.4594 | Yes | ||
| 149 | APPBP2 | 19944 | -5.768 | -0.4544 | Yes | ||
| 150 | DNMBP | 19947 | -5.770 | -0.4493 | Yes | ||
| 151 | PPP1R12A | 19974 | -5.824 | -0.4453 | Yes | ||
| 152 | RUNX1 | 20112 | -6.091 | -0.4461 | Yes | ||
| 153 | ATXN2 | 20152 | -6.172 | -0.4424 | Yes | ||
| 154 | SEC24B | 20156 | -6.176 | -0.4369 | Yes | ||
| 155 | TLK1 | 20203 | -6.276 | -0.4334 | Yes | ||
| 156 | GNAQ | 20205 | -6.283 | -0.4278 | Yes | ||
| 157 | WDR37 | 20224 | -6.332 | -0.4230 | Yes | ||
| 158 | ZCCHC11 | 20278 | -6.440 | -0.4196 | Yes | ||
| 159 | CNOT4 | 20347 | -6.588 | -0.4168 | Yes | ||
| 160 | STRN3 | 20390 | -6.698 | -0.4127 | Yes | ||
| 161 | PHF3 | 20412 | -6.763 | -0.4076 | Yes | ||
| 162 | ARHGEF7 | 20413 | -6.769 | -0.4015 | Yes | ||
| 163 | TIPARP | 20496 | -6.955 | -0.3991 | Yes | ||
| 164 | FBXW11 | 20554 | -7.084 | -0.3953 | Yes | ||
| 165 | TLE4 | 20591 | -7.180 | -0.3905 | Yes | ||
| 166 | DYRK1A | 20638 | -7.321 | -0.3861 | Yes | ||
| 167 | PTK2 | 20652 | -7.365 | -0.3800 | Yes | ||
| 168 | RNF13 | 20710 | -7.529 | -0.3759 | Yes | ||
| 169 | CHD1 | 20726 | -7.565 | -0.3698 | Yes | ||
| 170 | SETD2 | 20786 | -7.770 | -0.3655 | Yes | ||
| 171 | RBM16 | 20807 | -7.847 | -0.3594 | Yes | ||
| 172 | UBR2 | 20813 | -7.861 | -0.3526 | Yes | ||
| 173 | RBPSUH | 20825 | -7.896 | -0.3460 | Yes | ||
| 174 | RASA1 | 20833 | -7.927 | -0.3392 | Yes | ||
| 175 | MECP2 | 20899 | -8.166 | -0.3348 | Yes | ||
| 176 | RAI17 | 20912 | -8.217 | -0.3280 | Yes | ||
| 177 | PRKCBP1 | 20918 | -8.231 | -0.3208 | Yes | ||
| 178 | NCOA1 | 20923 | -8.265 | -0.3136 | Yes | ||
| 179 | SMURF2 | 20932 | -8.289 | -0.3065 | Yes | ||
| 180 | AKAP10 | 20957 | -8.399 | -0.3000 | Yes | ||
| 181 | AHDC1 | 20972 | -8.466 | -0.2931 | Yes | ||
| 182 | RBM39 | 21045 | -8.749 | -0.2885 | Yes | ||
| 183 | NUP153 | 21076 | -8.855 | -0.2820 | Yes | ||
| 184 | MYO10 | 21175 | -9.234 | -0.2782 | Yes | ||
| 185 | TSC22D2 | 21207 | -9.406 | -0.2711 | Yes | ||
| 186 | MYST3 | 21214 | -9.448 | -0.2629 | Yes | ||
| 187 | MYST4 | 21251 | -9.629 | -0.2559 | Yes | ||
| 188 | UGCG | 21296 | -9.876 | -0.2491 | Yes | ||
| 189 | FYN | 21301 | -9.911 | -0.2404 | Yes | ||
| 190 | AFF1 | 21312 | -9.956 | -0.2319 | Yes | ||
| 191 | HELZ | 21342 | -10.125 | -0.2241 | Yes | ||
| 192 | CDC2L5 | 21362 | -10.257 | -0.2158 | Yes | ||
| 193 | TMCC1 | 21366 | -10.274 | -0.2067 | Yes | ||
| 194 | CREBBP | 21411 | -10.605 | -0.1992 | Yes | ||
| 195 | SRPK2 | 21476 | -11.046 | -0.1922 | Yes | ||
| 196 | MTMR3 | 21499 | -11.309 | -0.1830 | Yes | ||
| 197 | NCOA3 | 21501 | -11.314 | -0.1729 | Yes | ||
| 198 | BAZ2B | 21508 | -11.364 | -0.1630 | Yes | ||
| 199 | CUTL1 | 21548 | -11.665 | -0.1543 | Yes | ||
| 200 | PAFAH1B1 | 21585 | -12.135 | -0.1450 | Yes | ||
| 201 | MARCH7 | 21597 | -12.301 | -0.1345 | Yes | ||
| 202 | TLK2 | 21642 | -12.867 | -0.1250 | Yes | ||
| 203 | SPEN | 21664 | -13.160 | -0.1141 | Yes | ||
| 204 | THRAP2 | 21740 | -14.503 | -0.1045 | Yes | ||
| 205 | IGF1R | 21784 | -15.295 | -0.0927 | Yes | ||
| 206 | CENTD1 | 21836 | -16.931 | -0.0799 | Yes | ||
| 207 | PCTK2 | 21857 | -17.748 | -0.0648 | Yes | ||
| 208 | MALT1 | 21871 | -18.066 | -0.0492 | Yes | ||
| 209 | NIPBL | 21877 | -18.673 | -0.0327 | Yes | ||
| 210 | TGFBR3 | 21891 | -19.317 | -0.0159 | Yes | ||
| 211 | KLF7 | 21904 | -20.512 | 0.0020 | Yes |