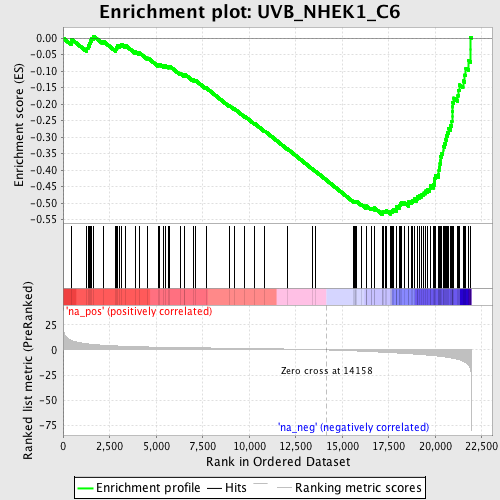
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | UVB_NHEK1_C6 |
| Enrichment Score (ES) | -0.5347472 |
| Normalized Enrichment Score (NES) | -2.1183004 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0017334494 |
| FWER p-Value | 0.028 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC7A1 | 445 | 9.366 | -0.0031 | No | ||
| 2 | RRS1 | 1280 | 6.096 | -0.0301 | No | ||
| 3 | XPO1 | 1342 | 5.926 | -0.0220 | No | ||
| 4 | CEBPD | 1427 | 5.760 | -0.0152 | No | ||
| 5 | BAZ1A | 1472 | 5.636 | -0.0069 | No | ||
| 6 | CUL1 | 1539 | 5.507 | 0.0003 | No | ||
| 7 | ORC2L | 1630 | 5.349 | 0.0060 | No | ||
| 8 | AUTS2 | 2152 | 4.610 | -0.0094 | No | ||
| 9 | CTBP2 | 2839 | 3.915 | -0.0336 | No | ||
| 10 | KIF23 | 2882 | 3.884 | -0.0284 | No | ||
| 11 | POLD3 | 2921 | 3.858 | -0.0230 | No | ||
| 12 | KIF11 | 3035 | 3.775 | -0.0212 | No | ||
| 13 | PLEKHC1 | 3143 | 3.699 | -0.0193 | No | ||
| 14 | POLE2 | 3339 | 3.542 | -0.0217 | No | ||
| 15 | KIF14 | 3898 | 3.202 | -0.0414 | No | ||
| 16 | MEIS1 | 4098 | 3.095 | -0.0448 | No | ||
| 17 | IRAK1BP1 | 4553 | 2.891 | -0.0603 | No | ||
| 18 | AJAP1 | 5103 | 2.685 | -0.0805 | No | ||
| 19 | TOP2A | 5189 | 2.657 | -0.0795 | No | ||
| 20 | HMGA2 | 5402 | 2.592 | -0.0844 | No | ||
| 21 | WWC1 | 5498 | 2.557 | -0.0840 | No | ||
| 22 | BCL7A | 5646 | 2.511 | -0.0862 | No | ||
| 23 | DOCK4 | 5715 | 2.491 | -0.0847 | No | ||
| 24 | VEGFC | 6286 | 2.307 | -0.1065 | No | ||
| 25 | MICAL2 | 6529 | 2.243 | -0.1135 | No | ||
| 26 | MTUS1 | 6548 | 2.239 | -0.1102 | No | ||
| 27 | SMAD3 | 6988 | 2.122 | -0.1264 | No | ||
| 28 | NFKB1 | 7091 | 2.093 | -0.1272 | No | ||
| 29 | GNE | 7677 | 1.937 | -0.1504 | No | ||
| 30 | MN1 | 8927 | 1.642 | -0.2046 | No | ||
| 31 | HDHD1A | 9205 | 1.574 | -0.2144 | No | ||
| 32 | CCNB2 | 9771 | 1.440 | -0.2376 | No | ||
| 33 | CENPC1 | 10306 | 1.310 | -0.2597 | No | ||
| 34 | ZZZ3 | 10839 | 1.178 | -0.2819 | No | ||
| 35 | CASP8 | 12035 | 0.830 | -0.3351 | No | ||
| 36 | NAV3 | 13380 | 0.365 | -0.3960 | No | ||
| 37 | MEGF9 | 13566 | 0.288 | -0.4039 | No | ||
| 38 | DOCK9 | 15619 | -0.954 | -0.4961 | No | ||
| 39 | IRS1 | 15669 | -0.991 | -0.4966 | No | ||
| 40 | FNBP1L | 15690 | -1.007 | -0.4956 | No | ||
| 41 | EIF2C2 | 15736 | -1.037 | -0.4958 | No | ||
| 42 | USP24 | 15777 | -1.065 | -0.4956 | No | ||
| 43 | DOPEY1 | 16029 | -1.264 | -0.5048 | No | ||
| 44 | SMC5 | 16277 | -1.465 | -0.5134 | No | ||
| 45 | LPHN2 | 16285 | -1.477 | -0.5110 | No | ||
| 46 | SLC25A36 | 16297 | -1.488 | -0.5088 | No | ||
| 47 | E2F5 | 16561 | -1.721 | -0.5177 | No | ||
| 48 | UBL3 | 16588 | -1.746 | -0.5156 | No | ||
| 49 | STRN | 16714 | -1.850 | -0.5180 | No | ||
| 50 | XPA | 16717 | -1.851 | -0.5146 | No | ||
| 51 | CDYL | 17157 | -2.270 | -0.5306 | Yes | ||
| 52 | PHLPP | 17173 | -2.284 | -0.5270 | Yes | ||
| 53 | ZNF638 | 17217 | -2.312 | -0.5248 | Yes | ||
| 54 | ATP13A3 | 17322 | -2.408 | -0.5251 | Yes | ||
| 55 | MLH3 | 17350 | -2.435 | -0.5218 | Yes | ||
| 56 | OTUD4 | 17607 | -2.691 | -0.5286 | Yes | ||
| 57 | CENPA | 17639 | -2.727 | -0.5250 | Yes | ||
| 58 | KLHL9 | 17696 | -2.780 | -0.5225 | Yes | ||
| 59 | FGFR2 | 17733 | -2.815 | -0.5189 | Yes | ||
| 60 | TBC1D8 | 17900 | -3.000 | -0.5210 | Yes | ||
| 61 | REV3L | 17923 | -3.021 | -0.5164 | Yes | ||
| 62 | PIP5K3 | 17927 | -3.024 | -0.5110 | Yes | ||
| 63 | TUBGCP3 | 18070 | -3.176 | -0.5117 | Yes | ||
| 64 | KIF2A | 18083 | -3.195 | -0.5063 | Yes | ||
| 65 | FRYL | 18121 | -3.241 | -0.5021 | Yes | ||
| 66 | ATP2B1 | 18161 | -3.285 | -0.4978 | Yes | ||
| 67 | PIP5K2B | 18340 | -3.463 | -0.4996 | Yes | ||
| 68 | EDD1 | 18562 | -3.718 | -0.5028 | Yes | ||
| 69 | RABGAP1L | 18576 | -3.732 | -0.4966 | Yes | ||
| 70 | ZHX2 | 18710 | -3.894 | -0.4955 | Yes | ||
| 71 | FNDC3A | 18769 | -3.963 | -0.4908 | Yes | ||
| 72 | PIK3C3 | 18859 | -4.098 | -0.4874 | Yes | ||
| 73 | CNOT2 | 19015 | -4.303 | -0.4865 | Yes | ||
| 74 | SLC9A6 | 19052 | -4.342 | -0.4802 | Yes | ||
| 75 | IGF2BP3 | 19161 | -4.519 | -0.4768 | Yes | ||
| 76 | WAPAL | 19275 | -4.690 | -0.4734 | Yes | ||
| 77 | ADCY9 | 19390 | -4.879 | -0.4696 | Yes | ||
| 78 | FOXO3A | 19458 | -4.980 | -0.4635 | Yes | ||
| 79 | BIRC2 | 19566 | -5.145 | -0.4589 | Yes | ||
| 80 | ATF2 | 19713 | -5.352 | -0.4557 | Yes | ||
| 81 | ABL1 | 19742 | -5.391 | -0.4471 | Yes | ||
| 82 | CREB3L2 | 19909 | -5.677 | -0.4442 | Yes | ||
| 83 | APPBP2 | 19944 | -5.768 | -0.4352 | Yes | ||
| 84 | PPP1R12A | 19974 | -5.824 | -0.4258 | Yes | ||
| 85 | NPAS2 | 19992 | -5.862 | -0.4158 | Yes | ||
| 86 | AYTL2 | 20172 | -6.218 | -0.4125 | Yes | ||
| 87 | SPOP | 20194 | -6.255 | -0.4020 | Yes | ||
| 88 | TLK1 | 20203 | -6.276 | -0.3908 | Yes | ||
| 89 | LSM14A | 20249 | -6.391 | -0.3811 | Yes | ||
| 90 | MAST4 | 20271 | -6.429 | -0.3702 | Yes | ||
| 91 | ZCCHC11 | 20278 | -6.440 | -0.3586 | Yes | ||
| 92 | CNOT4 | 20347 | -6.588 | -0.3496 | Yes | ||
| 93 | PHF3 | 20412 | -6.763 | -0.3400 | Yes | ||
| 94 | ARHGEF7 | 20413 | -6.769 | -0.3276 | Yes | ||
| 95 | SP3 | 20510 | -6.981 | -0.3191 | Yes | ||
| 96 | FOXO1A | 20562 | -7.105 | -0.3084 | Yes | ||
| 97 | TLE4 | 20591 | -7.180 | -0.2964 | Yes | ||
| 98 | PTK2 | 20652 | -7.365 | -0.2856 | Yes | ||
| 99 | RNF13 | 20710 | -7.529 | -0.2744 | Yes | ||
| 100 | RBPSUH | 20825 | -7.896 | -0.2650 | Yes | ||
| 101 | SLBP | 20885 | -8.119 | -0.2528 | Yes | ||
| 102 | MECP2 | 20899 | -8.166 | -0.2383 | Yes | ||
| 103 | SHOC2 | 20901 | -8.170 | -0.2233 | Yes | ||
| 104 | RAI17 | 20912 | -8.217 | -0.2087 | Yes | ||
| 105 | PRKCBP1 | 20918 | -8.231 | -0.1937 | Yes | ||
| 106 | CLASP1 | 20998 | -8.567 | -0.1816 | Yes | ||
| 107 | MYST3 | 21214 | -9.448 | -0.1740 | Yes | ||
| 108 | MYST4 | 21251 | -9.629 | -0.1579 | Yes | ||
| 109 | FYN | 21301 | -9.911 | -0.1419 | Yes | ||
| 110 | INSIG2 | 21489 | -11.220 | -0.1298 | Yes | ||
| 111 | PAFAH1B1 | 21585 | -12.135 | -0.1118 | Yes | ||
| 112 | TLK2 | 21642 | -12.867 | -0.0907 | Yes | ||
| 113 | SMC4 | 21798 | -15.658 | -0.0690 | Yes | ||
| 114 | KLF7 | 21904 | -20.512 | -0.0360 | Yes | ||
| 115 | SSBP2 | 21905 | -20.594 | 0.0019 | Yes |
