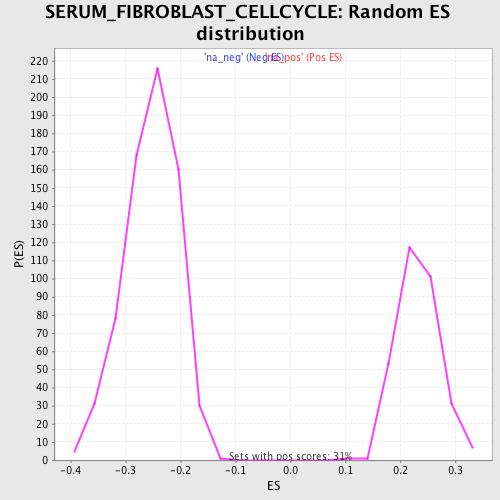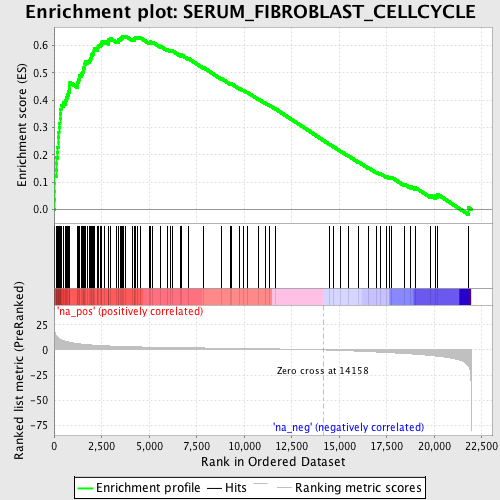
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | SERUM_FIBROBLAST_CELLCYCLE |
| Enrichment Score (ES) | 0.63457704 |
| Normalized Enrichment Score (NES) | 2.7280753 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SDC1 | 15 | 21.715 | 0.0347 | Yes | ||
| 2 | MCM6 | 26 | 19.246 | 0.0657 | Yes | ||
| 3 | AMD1 | 32 | 18.200 | 0.0951 | Yes | ||
| 4 | MND1 | 36 | 17.699 | 0.1239 | Yes | ||
| 5 | PRIM1 | 103 | 14.426 | 0.1444 | Yes | ||
| 6 | TIPIN | 111 | 14.254 | 0.1673 | Yes | ||
| 7 | LYAR | 117 | 14.016 | 0.1900 | Yes | ||
| 8 | WDR51A | 163 | 12.655 | 0.2085 | Yes | ||
| 9 | CKS1B | 182 | 12.344 | 0.2279 | Yes | ||
| 10 | GMNN | 209 | 11.840 | 0.2460 | Yes | ||
| 11 | CDCA5 | 211 | 11.784 | 0.2652 | Yes | ||
| 12 | EXO1 | 228 | 11.562 | 0.2833 | Yes | ||
| 13 | CDCA8 | 258 | 11.203 | 0.3002 | Yes | ||
| 14 | MCM4 | 293 | 10.788 | 0.3163 | Yes | ||
| 15 | DLG7 | 310 | 10.669 | 0.3330 | Yes | ||
| 16 | TPX2 | 318 | 10.605 | 0.3499 | Yes | ||
| 17 | TRIP13 | 337 | 10.432 | 0.3661 | Yes | ||
| 18 | PRIM2A | 370 | 10.094 | 0.3811 | Yes | ||
| 19 | CKS2 | 491 | 9.082 | 0.3905 | Yes | ||
| 20 | GTSE1 | 596 | 8.460 | 0.3995 | Yes | ||
| 21 | RRM1 | 643 | 8.235 | 0.4108 | Yes | ||
| 22 | RFC4 | 716 | 7.927 | 0.4205 | Yes | ||
| 23 | DHFR | 756 | 7.733 | 0.4313 | Yes | ||
| 24 | FEN1 | 812 | 7.526 | 0.4411 | Yes | ||
| 25 | FANCA | 823 | 7.494 | 0.4528 | Yes | ||
| 26 | AURKA | 833 | 7.454 | 0.4646 | Yes | ||
| 27 | MBOAT1 | 1227 | 6.220 | 0.4567 | Yes | ||
| 28 | TACC3 | 1239 | 6.178 | 0.4663 | Yes | ||
| 29 | RECQL4 | 1283 | 6.094 | 0.4743 | Yes | ||
| 30 | PTTG1 | 1313 | 6.003 | 0.4827 | Yes | ||
| 31 | MCM5 | 1329 | 5.965 | 0.4918 | Yes | ||
| 32 | CKAP2 | 1442 | 5.718 | 0.4960 | Yes | ||
| 33 | PWP1 | 1496 | 5.602 | 0.5027 | Yes | ||
| 34 | RAD51AP1 | 1524 | 5.559 | 0.5105 | Yes | ||
| 35 | NCAPH | 1557 | 5.482 | 0.5180 | Yes | ||
| 36 | SPAG5 | 1617 | 5.373 | 0.5241 | Yes | ||
| 37 | CDC25C | 1619 | 5.371 | 0.5328 | Yes | ||
| 38 | CDCA7 | 1636 | 5.336 | 0.5408 | Yes | ||
| 39 | USP1 | 1782 | 5.111 | 0.5425 | Yes | ||
| 40 | MAD2L1 | 1861 | 4.996 | 0.5470 | Yes | ||
| 41 | CDC6 | 1934 | 4.895 | 0.5517 | Yes | ||
| 42 | CDCA1 | 1956 | 4.866 | 0.5587 | Yes | ||
| 43 | RFC2 | 1973 | 4.841 | 0.5659 | Yes | ||
| 44 | EZH2 | 2009 | 4.793 | 0.5721 | Yes | ||
| 45 | LMNB1 | 2084 | 4.716 | 0.5764 | Yes | ||
| 46 | RANGAP1 | 2098 | 4.700 | 0.5835 | Yes | ||
| 47 | UHRF1 | 2130 | 4.635 | 0.5896 | Yes | ||
| 48 | CKAP2L | 2295 | 4.423 | 0.5893 | Yes | ||
| 49 | TUBB2C | 2308 | 4.412 | 0.5960 | Yes | ||
| 50 | CDC25A | 2359 | 4.348 | 0.6008 | Yes | ||
| 51 | PBK | 2430 | 4.285 | 0.6045 | Yes | ||
| 52 | PCNA | 2502 | 4.211 | 0.6082 | Yes | ||
| 53 | KIFC1 | 2519 | 4.191 | 0.6143 | Yes | ||
| 54 | KNTC1 | 2633 | 4.074 | 0.6157 | Yes | ||
| 55 | TIMP1 | 2871 | 3.893 | 0.6112 | Yes | ||
| 56 | ILF2 | 2878 | 3.887 | 0.6173 | Yes | ||
| 57 | KIF23 | 2882 | 3.884 | 0.6235 | Yes | ||
| 58 | HMMR | 2973 | 3.819 | 0.6256 | Yes | ||
| 59 | CCNA2 | 3305 | 3.575 | 0.6163 | Yes | ||
| 60 | CASP3 | 3387 | 3.501 | 0.6183 | Yes | ||
| 61 | HELLS | 3407 | 3.491 | 0.6231 | Yes | ||
| 62 | MELK | 3493 | 3.439 | 0.6248 | Yes | ||
| 63 | CCDC99 | 3545 | 3.409 | 0.6281 | Yes | ||
| 64 | FOXM1 | 3582 | 3.376 | 0.6319 | Yes | ||
| 65 | PLK1 | 3661 | 3.326 | 0.6338 | Yes | ||
| 66 | ANP32E | 3761 | 3.275 | 0.6346 | Yes | ||
| 67 | MLF1IP | 4109 | 3.092 | 0.6237 | No | ||
| 68 | ATAD2 | 4214 | 3.035 | 0.6239 | No | ||
| 69 | PAQR4 | 4246 | 3.021 | 0.6274 | No | ||
| 70 | UBE2C | 4277 | 3.007 | 0.6310 | No | ||
| 71 | ANLN | 4377 | 2.963 | 0.6313 | No | ||
| 72 | TUBB | 4527 | 2.901 | 0.6292 | No | ||
| 73 | PSRC1 | 5012 | 2.716 | 0.6114 | No | ||
| 74 | CDKN3 | 5062 | 2.699 | 0.6136 | No | ||
| 75 | TOP2A | 5189 | 2.657 | 0.6122 | No | ||
| 76 | MET | 5581 | 2.531 | 0.5984 | No | ||
| 77 | GINS3 | 5956 | 2.411 | 0.5852 | No | ||
| 78 | RRM2 | 6132 | 2.358 | 0.5810 | No | ||
| 79 | BUB1 | 6204 | 2.335 | 0.5816 | No | ||
| 80 | SGCD | 6671 | 2.203 | 0.5638 | No | ||
| 81 | MCM8 | 6691 | 2.199 | 0.5666 | No | ||
| 82 | CENPF | 7069 | 2.101 | 0.5527 | No | ||
| 83 | ASF1B | 7860 | 1.892 | 0.5196 | No | ||
| 84 | ESCO2 | 8820 | 1.668 | 0.4784 | No | ||
| 85 | MPHOSPH1 | 9264 | 1.561 | 0.4607 | No | ||
| 86 | DONSON | 9324 | 1.546 | 0.4605 | No | ||
| 87 | CCNB2 | 9771 | 1.440 | 0.4424 | No | ||
| 88 | CCNF | 9940 | 1.400 | 0.4370 | No | ||
| 89 | GAS2L3 | 10155 | 1.346 | 0.4294 | No | ||
| 90 | MAPK13 | 10765 | 1.197 | 0.4035 | No | ||
| 91 | PHTF2 | 11105 | 1.107 | 0.3898 | No | ||
| 92 | CIITA | 11335 | 1.045 | 0.3810 | No | ||
| 93 | BARD1 | 11636 | 0.955 | 0.3688 | No | ||
| 94 | FANCG | 14489 | -0.216 | 0.2386 | No | ||
| 95 | EFHC1 | 14714 | -0.352 | 0.2289 | No | ||
| 96 | UBE2T | 15083 | -0.596 | 0.2130 | No | ||
| 97 | ADAMTS1 | 15473 | -0.862 | 0.1966 | No | ||
| 98 | DEPDC1B | 16020 | -1.256 | 0.1736 | No | ||
| 99 | H2AFX | 16546 | -1.701 | 0.1524 | No | ||
| 100 | HN1 | 16966 | -2.091 | 0.1366 | No | ||
| 101 | ABCC5 | 17172 | -2.283 | 0.1309 | No | ||
| 102 | CTNNA1 | 17513 | -2.616 | 0.1196 | No | ||
| 103 | CENPA | 17639 | -2.727 | 0.1183 | No | ||
| 104 | TYMS | 17771 | -2.857 | 0.1170 | No | ||
| 105 | WSB1 | 18449 | -3.600 | 0.0919 | No | ||
| 106 | FBXL20 | 18750 | -3.942 | 0.0846 | No | ||
| 107 | ITGB3 | 19031 | -4.324 | 0.0788 | No | ||
| 108 | ANKRD10 | 19817 | -5.521 | 0.0519 | No | ||
| 109 | TUBA1 | 20075 | -6.030 | 0.0499 | No | ||
| 110 | YWHAH | 20180 | -6.225 | 0.0553 | No | ||
| 111 | SMC4 | 21798 | -15.658 | 0.0068 | No |