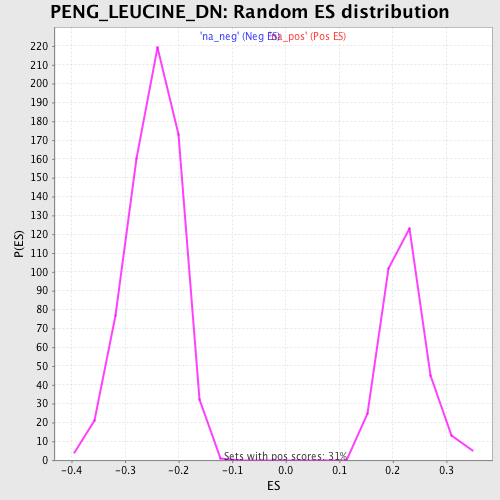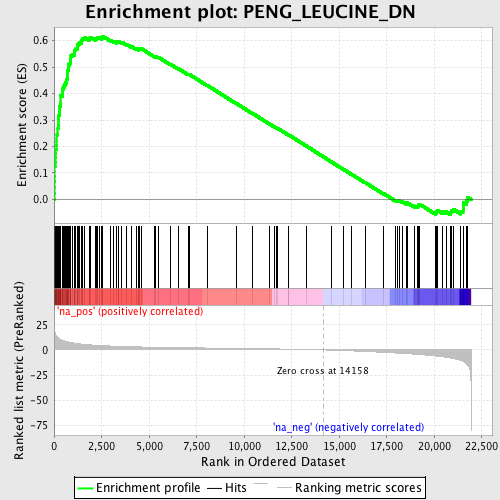
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | PENG_LEUCINE_DN |
| Enrichment Score (ES) | 0.61734265 |
| Normalized Enrichment Score (NES) | 2.7646575 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCT5 | 9 | 22.744 | 0.0241 | Yes | ||
| 2 | CAD | 18 | 21.326 | 0.0468 | Yes | ||
| 3 | SNRPA1 | 22 | 19.752 | 0.0679 | Yes | ||
| 4 | AMD1 | 32 | 18.200 | 0.0871 | Yes | ||
| 5 | CYCS | 42 | 17.128 | 0.1052 | Yes | ||
| 6 | TIMM17A | 44 | 16.991 | 0.1235 | Yes | ||
| 7 | GART | 50 | 16.738 | 0.1413 | Yes | ||
| 8 | LDHA | 59 | 16.261 | 0.1585 | Yes | ||
| 9 | DNAJA1 | 67 | 15.936 | 0.1754 | Yes | ||
| 10 | SLC29A1 | 88 | 14.935 | 0.1906 | Yes | ||
| 11 | NCL | 125 | 13.890 | 0.2039 | Yes | ||
| 12 | HPRT1 | 145 | 13.325 | 0.2174 | Yes | ||
| 13 | DDX21 | 148 | 13.171 | 0.2315 | Yes | ||
| 14 | PSMA7 | 149 | 13.085 | 0.2456 | Yes | ||
| 15 | PSMD14 | 200 | 11.991 | 0.2563 | Yes | ||
| 16 | CCT4 | 201 | 11.986 | 0.2692 | Yes | ||
| 17 | XRCC5 | 213 | 11.773 | 0.2814 | Yes | ||
| 18 | SQLE | 226 | 11.580 | 0.2933 | Yes | ||
| 19 | TCP1 | 230 | 11.552 | 0.3057 | Yes | ||
| 20 | DDX1 | 243 | 11.395 | 0.3174 | Yes | ||
| 21 | CCT6A | 274 | 11.054 | 0.3280 | Yes | ||
| 22 | MCM4 | 293 | 10.788 | 0.3388 | Yes | ||
| 23 | HSPE1 | 301 | 10.746 | 0.3500 | Yes | ||
| 24 | MRPL12 | 316 | 10.627 | 0.3609 | Yes | ||
| 25 | TRIP13 | 337 | 10.432 | 0.3712 | Yes | ||
| 26 | VDAC1 | 339 | 10.409 | 0.3824 | Yes | ||
| 27 | MTHFD1 | 340 | 10.406 | 0.3936 | Yes | ||
| 28 | UMPS | 438 | 9.445 | 0.3993 | Yes | ||
| 29 | PSMA5 | 455 | 9.295 | 0.4086 | Yes | ||
| 30 | MRPL28 | 459 | 9.283 | 0.4185 | Yes | ||
| 31 | PSMD1 | 496 | 9.051 | 0.4266 | Yes | ||
| 32 | HSPA4 | 539 | 8.755 | 0.4342 | Yes | ||
| 33 | SFRS2 | 597 | 8.452 | 0.4407 | Yes | ||
| 34 | MRPS12 | 666 | 8.123 | 0.4463 | Yes | ||
| 35 | PSMF1 | 671 | 8.108 | 0.4549 | Yes | ||
| 36 | SRM | 702 | 7.962 | 0.4621 | Yes | ||
| 37 | NMT1 | 709 | 7.940 | 0.4704 | Yes | ||
| 38 | TFRC | 712 | 7.936 | 0.4788 | Yes | ||
| 39 | ETF1 | 722 | 7.905 | 0.4870 | Yes | ||
| 40 | CCT2 | 755 | 7.735 | 0.4938 | Yes | ||
| 41 | CYC1 | 767 | 7.693 | 0.5016 | Yes | ||
| 42 | MRPL3 | 769 | 7.691 | 0.5099 | Yes | ||
| 43 | PSMB7 | 821 | 7.495 | 0.5156 | Yes | ||
| 44 | FASN | 864 | 7.363 | 0.5216 | Yes | ||
| 45 | NOL5A | 873 | 7.320 | 0.5292 | Yes | ||
| 46 | TOMM40 | 884 | 7.276 | 0.5366 | Yes | ||
| 47 | NME2 | 885 | 7.275 | 0.5444 | Yes | ||
| 48 | SSB | 963 | 6.987 | 0.5484 | Yes | ||
| 49 | ERH | 1082 | 6.622 | 0.5501 | Yes | ||
| 50 | UNG | 1088 | 6.592 | 0.5570 | Yes | ||
| 51 | TK1 | 1089 | 6.588 | 0.5641 | Yes | ||
| 52 | CSE1L | 1142 | 6.454 | 0.5687 | Yes | ||
| 53 | CCNB1 | 1213 | 6.264 | 0.5723 | Yes | ||
| 54 | ADRM1 | 1226 | 6.220 | 0.5784 | Yes | ||
| 55 | NOL1 | 1232 | 6.212 | 0.5849 | Yes | ||
| 56 | BST2 | 1274 | 6.105 | 0.5896 | Yes | ||
| 57 | CTSC | 1335 | 5.959 | 0.5933 | Yes | ||
| 58 | EI24 | 1424 | 5.764 | 0.5955 | Yes | ||
| 59 | FDPS | 1437 | 5.730 | 0.6011 | Yes | ||
| 60 | RNF126 | 1482 | 5.618 | 0.6051 | Yes | ||
| 61 | NUTF2 | 1511 | 5.580 | 0.6099 | Yes | ||
| 62 | PSMD3 | 1622 | 5.364 | 0.6106 | Yes | ||
| 63 | FKBP4 | 1842 | 5.023 | 0.6060 | Yes | ||
| 64 | ILF3 | 1868 | 4.988 | 0.6102 | Yes | ||
| 65 | BYSL | 1933 | 4.897 | 0.6126 | Yes | ||
| 66 | ALDOA | 2183 | 4.556 | 0.6061 | Yes | ||
| 67 | ME2 | 2220 | 4.521 | 0.6093 | Yes | ||
| 68 | EIF4G2 | 2275 | 4.447 | 0.6116 | Yes | ||
| 69 | FKBP2 | 2388 | 4.322 | 0.6112 | Yes | ||
| 70 | IDI1 | 2518 | 4.194 | 0.6098 | Yes | ||
| 71 | CDC20 | 2524 | 4.184 | 0.6141 | Yes | ||
| 72 | SEC61B | 2551 | 4.153 | 0.6173 | Yes | ||
| 73 | GGA2 | 2976 | 3.817 | 0.6020 | No | ||
| 74 | HSPA5 | 3144 | 3.699 | 0.5984 | No | ||
| 75 | CCNA2 | 3305 | 3.575 | 0.5949 | No | ||
| 76 | CALR | 3379 | 3.515 | 0.5953 | No | ||
| 77 | UBE2N | 3412 | 3.489 | 0.5976 | No | ||
| 78 | AHSA1 | 3558 | 3.393 | 0.5946 | No | ||
| 79 | ACAT2 | 3828 | 3.238 | 0.5858 | No | ||
| 80 | VRK1 | 4077 | 3.102 | 0.5778 | No | ||
| 81 | MAP2K3 | 4337 | 2.980 | 0.5691 | No | ||
| 82 | DHCR24 | 4440 | 2.936 | 0.5676 | No | ||
| 83 | EIF4G1 | 4441 | 2.936 | 0.5708 | No | ||
| 84 | TCEB1 | 4482 | 2.920 | 0.5721 | No | ||
| 85 | PTBP1 | 4573 | 2.885 | 0.5711 | No | ||
| 86 | DHX9 | 5300 | 2.623 | 0.5406 | No | ||
| 87 | ARMET | 5341 | 2.610 | 0.5416 | No | ||
| 88 | PFKM | 5492 | 2.559 | 0.5375 | No | ||
| 89 | DHRS1 | 6126 | 2.359 | 0.5110 | No | ||
| 90 | NBL1 | 6543 | 2.240 | 0.4944 | No | ||
| 91 | SCD | 7062 | 2.104 | 0.4729 | No | ||
| 92 | EBP | 7109 | 2.087 | 0.4730 | No | ||
| 93 | PIK3R3 | 8058 | 1.844 | 0.4316 | No | ||
| 94 | OGFR | 9576 | 1.488 | 0.3637 | No | ||
| 95 | NP | 10450 | 1.275 | 0.3250 | No | ||
| 96 | PPIB | 11321 | 1.049 | 0.2863 | No | ||
| 97 | COPB2 | 11623 | 0.959 | 0.2735 | No | ||
| 98 | PSMD2 | 11699 | 0.938 | 0.2711 | No | ||
| 99 | EIF2B4 | 11781 | 0.911 | 0.2683 | No | ||
| 100 | GCDH | 12329 | 0.746 | 0.2441 | No | ||
| 101 | EMP3 | 12355 | 0.738 | 0.2437 | No | ||
| 102 | MLF2 | 13305 | 0.391 | 0.2006 | No | ||
| 103 | ALG8 | 14580 | -0.265 | 0.1425 | No | ||
| 104 | RBM14 | 15224 | -0.692 | 0.1138 | No | ||
| 105 | EMD | 15672 | -0.995 | 0.0944 | No | ||
| 106 | MMP15 | 16362 | -1.541 | 0.0645 | No | ||
| 107 | PTPN6 | 17353 | -2.437 | 0.0217 | No | ||
| 108 | NXF1 | 17961 | -3.065 | -0.0028 | No | ||
| 109 | TOP1 | 18087 | -3.196 | -0.0051 | No | ||
| 110 | GLO1 | 18175 | -3.299 | -0.0055 | No | ||
| 111 | EIF5 | 18319 | -3.440 | -0.0084 | No | ||
| 112 | DNAJB6 | 18542 | -3.699 | -0.0145 | No | ||
| 113 | XPO6 | 18572 | -3.723 | -0.0119 | No | ||
| 114 | POLRMT | 18956 | -4.220 | -0.0249 | No | ||
| 115 | PDCD4 | 19102 | -4.419 | -0.0267 | No | ||
| 116 | UBC | 19151 | -4.504 | -0.0241 | No | ||
| 117 | PSMD11 | 19193 | -4.572 | -0.0210 | No | ||
| 118 | BRD2 | 19233 | -4.628 | -0.0178 | No | ||
| 119 | TUBA1 | 20075 | -6.030 | -0.0499 | No | ||
| 120 | NUP98 | 20141 | -6.156 | -0.0462 | No | ||
| 121 | PAPOLA | 20154 | -6.173 | -0.0401 | No | ||
| 122 | LIMK2 | 20416 | -6.771 | -0.0448 | No | ||
| 123 | DYRK1A | 20638 | -7.321 | -0.0470 | No | ||
| 124 | SPINT2 | 20857 | -8.002 | -0.0483 | No | ||
| 125 | TGFB1 | 20897 | -8.163 | -0.0413 | No | ||
| 126 | POLG | 21020 | -8.661 | -0.0376 | No | ||
| 127 | WDR1 | 21407 | -10.582 | -0.0439 | No | ||
| 128 | RASSF2 | 21521 | -11.453 | -0.0367 | No | ||
| 129 | GSPT1 | 21530 | -11.532 | -0.0246 | No | ||
| 130 | LDHB | 21541 | -11.617 | -0.0125 | No | ||
| 131 | TNFRSF1A | 21703 | -13.816 | -0.0050 | No | ||
| 132 | PSME4 | 21771 | -14.970 | 0.0081 | No |