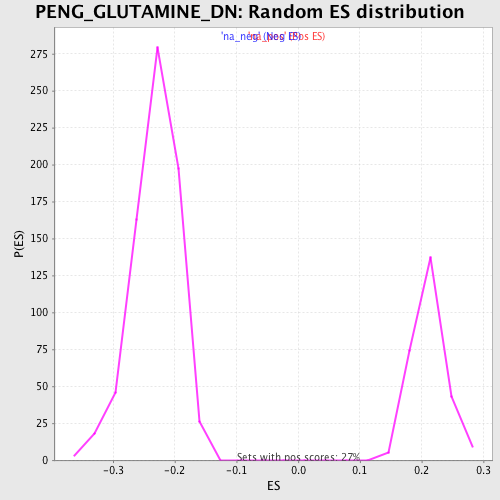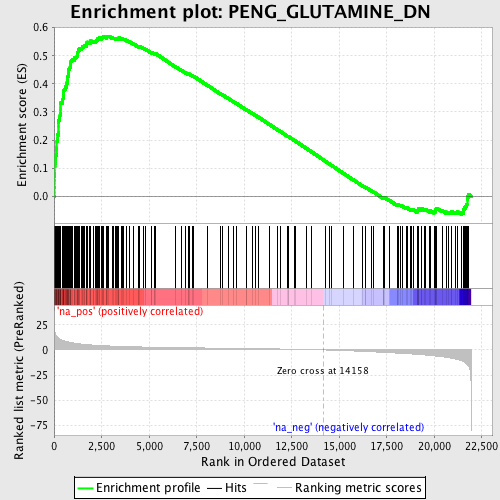
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | PENG_GLUTAMINE_DN |
| Enrichment Score (ES) | 0.56985986 |
| Normalized Enrichment Score (NES) | 2.7087114 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCT5 | 9 | 22.744 | 0.0146 | Yes | ||
| 2 | C1QBP | 10 | 22.644 | 0.0296 | Yes | ||
| 3 | CAD | 18 | 21.326 | 0.0434 | Yes | ||
| 4 | FBL | 19 | 21.204 | 0.0575 | Yes | ||
| 5 | SNRPA1 | 22 | 19.752 | 0.0704 | Yes | ||
| 6 | AMD1 | 32 | 18.200 | 0.0821 | Yes | ||
| 7 | CYCS | 42 | 17.128 | 0.0930 | Yes | ||
| 8 | TIMM17A | 44 | 16.991 | 0.1042 | Yes | ||
| 9 | PA2G4 | 48 | 16.873 | 0.1152 | Yes | ||
| 10 | LDHA | 59 | 16.261 | 0.1255 | Yes | ||
| 11 | DNAJA1 | 67 | 15.936 | 0.1357 | Yes | ||
| 12 | SLC29A1 | 88 | 14.935 | 0.1447 | Yes | ||
| 13 | PES1 | 120 | 13.982 | 0.1525 | Yes | ||
| 14 | NCL | 125 | 13.890 | 0.1616 | Yes | ||
| 15 | PHB | 132 | 13.657 | 0.1703 | Yes | ||
| 16 | HPRT1 | 145 | 13.325 | 0.1786 | Yes | ||
| 17 | HNRPAB | 147 | 13.193 | 0.1873 | Yes | ||
| 18 | DDX21 | 148 | 13.171 | 0.1960 | Yes | ||
| 19 | IFRD2 | 185 | 12.297 | 0.2025 | Yes | ||
| 20 | PSMB3 | 190 | 12.244 | 0.2104 | Yes | ||
| 21 | PRDX4 | 193 | 12.146 | 0.2183 | Yes | ||
| 22 | XRCC5 | 213 | 11.773 | 0.2253 | Yes | ||
| 23 | GMPS | 224 | 11.600 | 0.2325 | Yes | ||
| 24 | NOLC1 | 225 | 11.590 | 0.2401 | Yes | ||
| 25 | SQLE | 226 | 11.580 | 0.2478 | Yes | ||
| 26 | EIF5A | 229 | 11.555 | 0.2554 | Yes | ||
| 27 | TCP1 | 230 | 11.552 | 0.2630 | Yes | ||
| 28 | DDX1 | 243 | 11.395 | 0.2700 | Yes | ||
| 29 | CCT6A | 274 | 11.054 | 0.2759 | Yes | ||
| 30 | HSPE1 | 301 | 10.746 | 0.2818 | Yes | ||
| 31 | EBNA1BP2 | 307 | 10.700 | 0.2887 | Yes | ||
| 32 | MRPL12 | 316 | 10.627 | 0.2954 | Yes | ||
| 33 | CTPS | 332 | 10.485 | 0.3016 | Yes | ||
| 34 | RQCD1 | 333 | 10.476 | 0.3085 | Yes | ||
| 35 | TRIP13 | 337 | 10.432 | 0.3153 | Yes | ||
| 36 | VDAC1 | 339 | 10.409 | 0.3222 | Yes | ||
| 37 | MTHFD1 | 340 | 10.406 | 0.3290 | Yes | ||
| 38 | RNU3IP2 | 358 | 10.194 | 0.3350 | Yes | ||
| 39 | UMPS | 438 | 9.445 | 0.3376 | Yes | ||
| 40 | MRPL28 | 459 | 9.283 | 0.3428 | Yes | ||
| 41 | RUVBL2 | 467 | 9.229 | 0.3486 | Yes | ||
| 42 | MAPRE2 | 476 | 9.183 | 0.3543 | Yes | ||
| 43 | PSMD12 | 484 | 9.145 | 0.3601 | Yes | ||
| 44 | SNRPF | 492 | 9.081 | 0.3658 | Yes | ||
| 45 | PSMD1 | 496 | 9.051 | 0.3716 | Yes | ||
| 46 | HRAS | 510 | 8.966 | 0.3770 | Yes | ||
| 47 | HSPA4 | 539 | 8.755 | 0.3815 | Yes | ||
| 48 | COPS3 | 584 | 8.501 | 0.3851 | Yes | ||
| 49 | SFRS2 | 597 | 8.452 | 0.3901 | Yes | ||
| 50 | PSMB6 | 642 | 8.237 | 0.3935 | Yes | ||
| 51 | MRPS12 | 666 | 8.123 | 0.3978 | Yes | ||
| 52 | PSMF1 | 671 | 8.108 | 0.4030 | Yes | ||
| 53 | SLC5A6 | 694 | 7.993 | 0.4073 | Yes | ||
| 54 | SRM | 702 | 7.962 | 0.4122 | Yes | ||
| 55 | NMT1 | 709 | 7.940 | 0.4172 | Yes | ||
| 56 | IDH3A | 711 | 7.936 | 0.4224 | Yes | ||
| 57 | TFRC | 712 | 7.936 | 0.4277 | Yes | ||
| 58 | PAICS | 742 | 7.803 | 0.4315 | Yes | ||
| 59 | CCT2 | 755 | 7.735 | 0.4361 | Yes | ||
| 60 | PRPS1 | 761 | 7.714 | 0.4410 | Yes | ||
| 61 | CYC1 | 767 | 7.693 | 0.4458 | Yes | ||
| 62 | MRPL3 | 769 | 7.691 | 0.4509 | Yes | ||
| 63 | NUDC | 799 | 7.598 | 0.4546 | Yes | ||
| 64 | SYNGR2 | 834 | 7.454 | 0.4579 | Yes | ||
| 65 | FASN | 864 | 7.363 | 0.4615 | Yes | ||
| 66 | SYNCRIP | 870 | 7.330 | 0.4661 | Yes | ||
| 67 | NOL5A | 873 | 7.320 | 0.4708 | Yes | ||
| 68 | NME1 | 883 | 7.284 | 0.4752 | Yes | ||
| 69 | TOMM40 | 884 | 7.276 | 0.4801 | Yes | ||
| 70 | MORF4L2 | 917 | 7.150 | 0.4833 | Yes | ||
| 71 | SSB | 963 | 6.987 | 0.4859 | Yes | ||
| 72 | METTL1 | 1062 | 6.693 | 0.4858 | Yes | ||
| 73 | ERH | 1082 | 6.622 | 0.4893 | Yes | ||
| 74 | PSMA6 | 1086 | 6.599 | 0.4935 | Yes | ||
| 75 | CSE1L | 1142 | 6.454 | 0.4953 | Yes | ||
| 76 | POLR2C | 1202 | 6.285 | 0.4967 | Yes | ||
| 77 | EIF2B5 | 1212 | 6.265 | 0.5004 | Yes | ||
| 78 | CCNB1 | 1213 | 6.264 | 0.5046 | Yes | ||
| 79 | ADRM1 | 1226 | 6.220 | 0.5082 | Yes | ||
| 80 | NOL1 | 1232 | 6.212 | 0.5120 | Yes | ||
| 81 | RPS26 | 1258 | 6.138 | 0.5149 | Yes | ||
| 82 | RRS1 | 1280 | 6.096 | 0.5180 | Yes | ||
| 83 | PPIF | 1291 | 6.057 | 0.5216 | Yes | ||
| 84 | CTSC | 1335 | 5.959 | 0.5235 | Yes | ||
| 85 | XPO1 | 1342 | 5.926 | 0.5272 | Yes | ||
| 86 | PIK3R2 | 1448 | 5.700 | 0.5261 | Yes | ||
| 87 | BAZ1A | 1472 | 5.636 | 0.5288 | Yes | ||
| 88 | RNF126 | 1482 | 5.618 | 0.5321 | Yes | ||
| 89 | NDUFS6 | 1564 | 5.471 | 0.5320 | Yes | ||
| 90 | SSBP1 | 1577 | 5.457 | 0.5350 | Yes | ||
| 91 | PSMD3 | 1622 | 5.364 | 0.5366 | Yes | ||
| 92 | SMARCC1 | 1696 | 5.252 | 0.5367 | Yes | ||
| 93 | AK2 | 1717 | 5.217 | 0.5392 | Yes | ||
| 94 | BOP1 | 1720 | 5.215 | 0.5426 | Yes | ||
| 95 | PRDX1 | 1726 | 5.201 | 0.5458 | Yes | ||
| 96 | ABCF1 | 1733 | 5.192 | 0.5489 | Yes | ||
| 97 | FKBP4 | 1842 | 5.023 | 0.5473 | Yes | ||
| 98 | TBL3 | 1919 | 4.913 | 0.5470 | Yes | ||
| 99 | BYSL | 1933 | 4.897 | 0.5497 | Yes | ||
| 100 | ATP2A2 | 1935 | 4.894 | 0.5529 | Yes | ||
| 101 | LMNB1 | 2084 | 4.716 | 0.5492 | Yes | ||
| 102 | RANGAP1 | 2098 | 4.700 | 0.5517 | Yes | ||
| 103 | ALDOA | 2183 | 4.556 | 0.5508 | Yes | ||
| 104 | MRPL23 | 2213 | 4.530 | 0.5525 | Yes | ||
| 105 | ME2 | 2220 | 4.521 | 0.5552 | Yes | ||
| 106 | EIF4G2 | 2275 | 4.447 | 0.5557 | Yes | ||
| 107 | PSMD8 | 2299 | 4.418 | 0.5575 | Yes | ||
| 108 | DDX48 | 2302 | 4.416 | 0.5604 | Yes | ||
| 109 | CDC25A | 2359 | 4.348 | 0.5607 | Yes | ||
| 110 | TUBG1 | 2365 | 4.341 | 0.5633 | Yes | ||
| 111 | FKBP2 | 2388 | 4.322 | 0.5652 | Yes | ||
| 112 | IDI1 | 2518 | 4.194 | 0.5620 | Yes | ||
| 113 | CDC20 | 2524 | 4.184 | 0.5645 | Yes | ||
| 114 | SEC61B | 2551 | 4.153 | 0.5661 | Yes | ||
| 115 | SLC19A1 | 2582 | 4.123 | 0.5674 | Yes | ||
| 116 | BZW1 | 2606 | 4.102 | 0.5691 | Yes | ||
| 117 | EEF1E1 | 2752 | 3.981 | 0.5650 | Yes | ||
| 118 | SNRPB2 | 2757 | 3.978 | 0.5675 | Yes | ||
| 119 | ATP5G3 | 2767 | 3.974 | 0.5697 | Yes | ||
| 120 | DDX18 | 2833 | 3.917 | 0.5693 | Yes | ||
| 121 | ILF2 | 2878 | 3.887 | 0.5699 | Yes | ||
| 122 | DHX15 | 3055 | 3.762 | 0.5642 | No | ||
| 123 | HSPA5 | 3144 | 3.699 | 0.5626 | No | ||
| 124 | HNRPA2B1 | 3240 | 3.624 | 0.5607 | No | ||
| 125 | RABGGTB | 3303 | 3.577 | 0.5602 | No | ||
| 126 | SLC29A2 | 3336 | 3.545 | 0.5610 | No | ||
| 127 | CALR | 3379 | 3.515 | 0.5614 | No | ||
| 128 | NUP160 | 3408 | 3.491 | 0.5625 | No | ||
| 129 | UBE2N | 3412 | 3.489 | 0.5646 | No | ||
| 130 | AHSA1 | 3558 | 3.393 | 0.5602 | No | ||
| 131 | SFRS1 | 3608 | 3.355 | 0.5602 | No | ||
| 132 | ALG3 | 3671 | 3.323 | 0.5595 | No | ||
| 133 | ACAT2 | 3828 | 3.238 | 0.5545 | No | ||
| 134 | BTK | 3984 | 3.157 | 0.5494 | No | ||
| 135 | MPHOSPH10 | 4191 | 3.050 | 0.5420 | No | ||
| 136 | DHCR24 | 4440 | 2.936 | 0.5325 | No | ||
| 137 | EIF4G1 | 4441 | 2.936 | 0.5344 | No | ||
| 138 | TCEB1 | 4482 | 2.920 | 0.5345 | No | ||
| 139 | ATR | 4687 | 2.841 | 0.5270 | No | ||
| 140 | ERF | 4798 | 2.793 | 0.5238 | No | ||
| 141 | PWP2H | 5122 | 2.680 | 0.5107 | No | ||
| 142 | SURF5 | 5126 | 2.678 | 0.5123 | No | ||
| 143 | BCAP31 | 5267 | 2.634 | 0.5076 | No | ||
| 144 | DHX9 | 5300 | 2.623 | 0.5079 | No | ||
| 145 | ARMET | 5341 | 2.610 | 0.5078 | No | ||
| 146 | PRPF4 | 6412 | 2.275 | 0.4600 | No | ||
| 147 | BMS1L | 6719 | 2.193 | 0.4473 | No | ||
| 148 | UBE2M | 6933 | 2.133 | 0.4389 | No | ||
| 149 | SCD | 7062 | 2.104 | 0.4344 | No | ||
| 150 | PTDSS1 | 7087 | 2.093 | 0.4347 | No | ||
| 151 | EBP | 7109 | 2.087 | 0.4351 | No | ||
| 152 | SRPK1 | 7263 | 2.046 | 0.4294 | No | ||
| 153 | CCND1 | 7333 | 2.028 | 0.4276 | No | ||
| 154 | PIK3R3 | 8058 | 1.844 | 0.3955 | No | ||
| 155 | LAMB3 | 8731 | 1.684 | 0.3657 | No | ||
| 156 | INHBC | 8864 | 1.657 | 0.3607 | No | ||
| 157 | UBE2G2 | 9154 | 1.587 | 0.3484 | No | ||
| 158 | KHSRP | 9423 | 1.524 | 0.3371 | No | ||
| 159 | OGFR | 9576 | 1.488 | 0.3311 | No | ||
| 160 | ADAM19 | 10140 | 1.349 | 0.3060 | No | ||
| 161 | IFI30 | 10142 | 1.349 | 0.3069 | No | ||
| 162 | NP | 10450 | 1.275 | 0.2936 | No | ||
| 163 | DOK4 | 10614 | 1.234 | 0.2869 | No | ||
| 164 | RNASEH1 | 10737 | 1.206 | 0.2821 | No | ||
| 165 | PPIB | 11321 | 1.049 | 0.2559 | No | ||
| 166 | EIF2B4 | 11781 | 0.911 | 0.2354 | No | ||
| 167 | GJB1 | 11888 | 0.875 | 0.2311 | No | ||
| 168 | GRIN2A | 12267 | 0.763 | 0.2142 | No | ||
| 169 | GCDH | 12329 | 0.746 | 0.2119 | No | ||
| 170 | EMP3 | 12355 | 0.738 | 0.2112 | No | ||
| 171 | RPLP2 | 12669 | 0.637 | 0.1972 | No | ||
| 172 | ATP1B3 | 12723 | 0.621 | 0.1952 | No | ||
| 173 | MLF2 | 13305 | 0.391 | 0.1687 | No | ||
| 174 | SGTA | 13522 | 0.307 | 0.1589 | No | ||
| 175 | MYD88 | 14281 | -0.077 | 0.1241 | No | ||
| 176 | PCBP1 | 14495 | -0.218 | 0.1144 | No | ||
| 177 | ALG8 | 14580 | -0.265 | 0.1107 | No | ||
| 178 | RBM14 | 15224 | -0.692 | 0.0816 | No | ||
| 179 | ZMPSTE24 | 15773 | -1.062 | 0.0570 | No | ||
| 180 | EWSR1 | 16252 | -1.451 | 0.0360 | No | ||
| 181 | MMP15 | 16362 | -1.541 | 0.0320 | No | ||
| 182 | MYST1 | 16412 | -1.584 | 0.0308 | No | ||
| 183 | ACLY | 16699 | -1.839 | 0.0188 | No | ||
| 184 | ARS2 | 16815 | -1.942 | 0.0148 | No | ||
| 185 | SC4MOL | 17325 | -2.413 | -0.0070 | No | ||
| 186 | GOLGA1 | 17349 | -2.435 | -0.0065 | No | ||
| 187 | PTPN6 | 17353 | -2.437 | -0.0050 | No | ||
| 188 | HNRPL | 17404 | -2.490 | -0.0057 | No | ||
| 189 | IDH1 | 17638 | -2.727 | -0.0146 | No | ||
| 190 | PFKP | 18081 | -3.193 | -0.0328 | No | ||
| 191 | TOP1 | 18087 | -3.196 | -0.0310 | No | ||
| 192 | TARDBP | 18107 | -3.229 | -0.0297 | No | ||
| 193 | RBBP6 | 18221 | -3.350 | -0.0327 | No | ||
| 194 | EIF5 | 18319 | -3.440 | -0.0349 | No | ||
| 195 | DNAJB6 | 18542 | -3.699 | -0.0426 | No | ||
| 196 | MAZ | 18575 | -3.730 | -0.0416 | No | ||
| 197 | ADCY3 | 18772 | -3.965 | -0.0481 | No | ||
| 198 | TAP1 | 18804 | -4.005 | -0.0468 | No | ||
| 199 | IMPDH1 | 18893 | -4.139 | -0.0481 | No | ||
| 200 | PDCD4 | 19102 | -4.419 | -0.0548 | No | ||
| 201 | DHCR7 | 19115 | -4.448 | -0.0524 | No | ||
| 202 | SF1 | 19149 | -4.500 | -0.0509 | No | ||
| 203 | SACS | 19160 | -4.518 | -0.0484 | No | ||
| 204 | PPP2R2A | 19189 | -4.568 | -0.0467 | No | ||
| 205 | PSMD11 | 19193 | -4.572 | -0.0438 | No | ||
| 206 | CLIC1 | 19322 | -4.768 | -0.0465 | No | ||
| 207 | MAP3K11 | 19341 | -4.796 | -0.0442 | No | ||
| 208 | FDFT1 | 19478 | -5.003 | -0.0471 | No | ||
| 209 | PPP1R10 | 19522 | -5.073 | -0.0458 | No | ||
| 210 | CELSR2 | 19775 | -5.441 | -0.0538 | No | ||
| 211 | CHD4 | 19790 | -5.476 | -0.0508 | No | ||
| 212 | PFN1 | 19993 | -5.863 | -0.0562 | No | ||
| 213 | TXNIP | 20050 | -5.974 | -0.0548 | No | ||
| 214 | SLC4A7 | 20059 | -6.000 | -0.0512 | No | ||
| 215 | TUBA1 | 20075 | -6.030 | -0.0479 | No | ||
| 216 | COG2 | 20098 | -6.071 | -0.0449 | No | ||
| 217 | NUP98 | 20141 | -6.156 | -0.0428 | No | ||
| 218 | LIMK2 | 20416 | -6.771 | -0.0509 | No | ||
| 219 | DYRK1A | 20638 | -7.321 | -0.0563 | No | ||
| 220 | FUS | 20756 | -7.663 | -0.0566 | No | ||
| 221 | TGFB1 | 20897 | -8.163 | -0.0576 | No | ||
| 222 | PRKCBP1 | 20918 | -8.231 | -0.0531 | No | ||
| 223 | SRRM1 | 21127 | -9.050 | -0.0567 | No | ||
| 224 | DNAJB1 | 21222 | -9.493 | -0.0547 | No | ||
| 225 | SFPQ | 21450 | -10.874 | -0.0580 | No | ||
| 226 | GSPT1 | 21530 | -11.532 | -0.0540 | No | ||
| 227 | LDHB | 21541 | -11.617 | -0.0468 | No | ||
| 228 | PABPN1 | 21586 | -12.140 | -0.0407 | No | ||
| 229 | STAT1 | 21656 | -13.091 | -0.0353 | No | ||
| 230 | HS6ST1 | 21698 | -13.762 | -0.0280 | No | ||
| 231 | YPEL1 | 21735 | -14.366 | -0.0202 | No | ||
| 232 | SNRP70 | 21736 | -14.371 | -0.0107 | No | ||
| 233 | PSME4 | 21771 | -14.970 | -0.0023 | No | ||
| 234 | XBP1 | 21801 | -15.697 | 0.0067 | No |