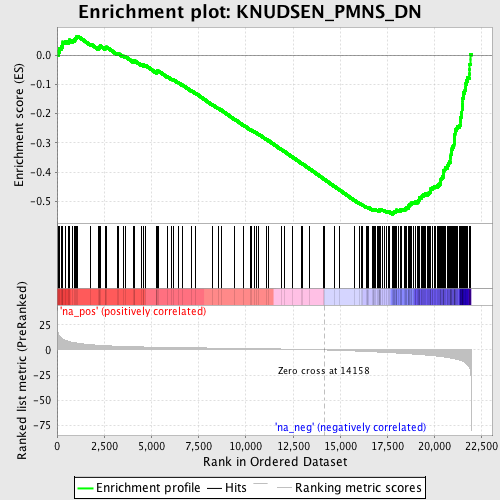
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | KNUDSEN_PMNS_DN |
| Enrichment Score (ES) | -0.543943 |
| Normalized Enrichment Score (NES) | -2.328684 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | COPS5 | 58 | 16.326 | 0.0123 | No | ||
| 2 | TKT | 124 | 13.910 | 0.0221 | No | ||
| 3 | XRCC5 | 213 | 11.773 | 0.0289 | No | ||
| 4 | COX5B | 277 | 11.008 | 0.0361 | No | ||
| 5 | SET | 308 | 10.690 | 0.0445 | No | ||
| 6 | NCF4 | 458 | 9.287 | 0.0462 | No | ||
| 7 | SLC25A3 | 603 | 8.417 | 0.0473 | No | ||
| 8 | TOPBP1 | 674 | 8.102 | 0.0515 | No | ||
| 9 | TRIM27 | 837 | 7.440 | 0.0509 | No | ||
| 10 | UQCR | 932 | 7.096 | 0.0531 | No | ||
| 11 | POLR2J | 974 | 6.950 | 0.0576 | No | ||
| 12 | PLCG2 | 1020 | 6.821 | 0.0618 | No | ||
| 13 | SMC1A | 1092 | 6.573 | 0.0645 | No | ||
| 14 | USP1 | 1782 | 5.111 | 0.0376 | No | ||
| 15 | TGFA | 2218 | 4.525 | 0.0217 | No | ||
| 16 | ME2 | 2220 | 4.521 | 0.0258 | No | ||
| 17 | CAMK2G | 2267 | 4.454 | 0.0278 | No | ||
| 18 | EIF4G2 | 2275 | 4.447 | 0.0315 | No | ||
| 19 | GAS7 | 2539 | 4.162 | 0.0233 | No | ||
| 20 | SLC19A1 | 2582 | 4.123 | 0.0251 | No | ||
| 21 | PRDX3 | 2612 | 4.097 | 0.0276 | No | ||
| 22 | TNPO3 | 3184 | 3.670 | 0.0047 | No | ||
| 23 | ANXA11 | 3233 | 3.631 | 0.0058 | No | ||
| 24 | UBE1C | 3499 | 3.436 | -0.0032 | No | ||
| 25 | SFRS1 | 3608 | 3.355 | -0.0051 | No | ||
| 26 | MYCBP | 4062 | 3.110 | -0.0231 | No | ||
| 27 | PNN | 4105 | 3.093 | -0.0222 | No | ||
| 28 | BARX2 | 4108 | 3.092 | -0.0194 | No | ||
| 29 | HTATIP2 | 4497 | 2.915 | -0.0346 | No | ||
| 30 | HHEX | 4574 | 2.884 | -0.0355 | No | ||
| 31 | ZFP36L2 | 4583 | 2.881 | -0.0332 | No | ||
| 32 | GNAO1 | 4698 | 2.837 | -0.0358 | No | ||
| 33 | RPS6KA1 | 5264 | 2.635 | -0.0594 | No | ||
| 34 | RIPK1 | 5284 | 2.628 | -0.0578 | No | ||
| 35 | TUBA2 | 5287 | 2.627 | -0.0555 | No | ||
| 36 | DHX9 | 5300 | 2.623 | -0.0537 | No | ||
| 37 | GNAT2 | 5369 | 2.603 | -0.0544 | No | ||
| 38 | CD46 | 5843 | 2.448 | -0.0739 | No | ||
| 39 | CITED2 | 6065 | 2.376 | -0.0819 | No | ||
| 40 | HSBP1 | 6146 | 2.353 | -0.0834 | No | ||
| 41 | ADAM10 | 6405 | 2.277 | -0.0932 | No | ||
| 42 | SEC14L1 | 6643 | 2.211 | -0.1020 | No | ||
| 43 | TCAP | 7126 | 2.083 | -0.1223 | No | ||
| 44 | RPS6KA5 | 7337 | 2.027 | -0.1301 | No | ||
| 45 | PIGB | 8242 | 1.802 | -0.1700 | No | ||
| 46 | SLC31A2 | 8534 | 1.735 | -0.1818 | No | ||
| 47 | TLR1 | 8686 | 1.694 | -0.1872 | No | ||
| 48 | TLR6 | 9407 | 1.528 | -0.2189 | No | ||
| 49 | GPR19 | 9899 | 1.410 | -0.2402 | No | ||
| 50 | RALY | 10271 | 1.318 | -0.2560 | No | ||
| 51 | SRP9 | 10291 | 1.314 | -0.2557 | No | ||
| 52 | CALM1 | 10454 | 1.273 | -0.2620 | No | ||
| 53 | GCA | 10542 | 1.250 | -0.2648 | No | ||
| 54 | DAPK2 | 10688 | 1.217 | -0.2704 | No | ||
| 55 | PDE3B | 11073 | 1.116 | -0.2870 | No | ||
| 56 | CSF3R | 11217 | 1.081 | -0.2926 | No | ||
| 57 | CALM2 | 11871 | 0.882 | -0.3218 | No | ||
| 58 | CASP8 | 12035 | 0.830 | -0.3286 | No | ||
| 59 | GALNT3 | 12486 | 0.697 | -0.3486 | No | ||
| 60 | MLLT7 | 12965 | 0.528 | -0.3701 | No | ||
| 61 | VIM | 13001 | 0.512 | -0.3712 | No | ||
| 62 | IL8RB | 13347 | 0.377 | -0.3868 | No | ||
| 63 | NFE2 | 14115 | 0.017 | -0.4220 | No | ||
| 64 | LST1 | 14171 | -0.014 | -0.4245 | No | ||
| 65 | TST | 14683 | -0.331 | -0.4477 | No | ||
| 66 | FEZ2 | 14963 | -0.507 | -0.4601 | No | ||
| 67 | ZMPSTE24 | 15773 | -1.062 | -0.4963 | No | ||
| 68 | CSTA | 16006 | -1.247 | -0.5058 | No | ||
| 69 | ARPC3 | 16106 | -1.329 | -0.5092 | No | ||
| 70 | DDX23 | 16204 | -1.409 | -0.5123 | No | ||
| 71 | GTF2E1 | 16385 | -1.558 | -0.5192 | No | ||
| 72 | CBX1 | 16443 | -1.607 | -0.5203 | No | ||
| 73 | XPC | 16452 | -1.614 | -0.5192 | No | ||
| 74 | CPNE3 | 16523 | -1.676 | -0.5209 | No | ||
| 75 | ACLY | 16699 | -1.839 | -0.5273 | No | ||
| 76 | GNB2 | 16743 | -1.876 | -0.5275 | No | ||
| 77 | CRADD | 16775 | -1.909 | -0.5272 | No | ||
| 78 | MBD4 | 16831 | -1.955 | -0.5279 | No | ||
| 79 | CD97 | 16847 | -1.975 | -0.5268 | No | ||
| 80 | SNAP23 | 16952 | -2.076 | -0.5297 | No | ||
| 81 | HCLS1 | 17013 | -2.142 | -0.5305 | No | ||
| 82 | AKT1 | 17066 | -2.193 | -0.5308 | No | ||
| 83 | OAZ1 | 17071 | -2.196 | -0.5290 | No | ||
| 84 | TYK2 | 17095 | -2.213 | -0.5280 | No | ||
| 85 | RGS19 | 17159 | -2.271 | -0.5288 | No | ||
| 86 | TROVE2 | 17160 | -2.272 | -0.5268 | No | ||
| 87 | RBMS1 | 17267 | -2.356 | -0.5295 | No | ||
| 88 | MAPKAPK3 | 17357 | -2.443 | -0.5313 | No | ||
| 89 | UPF1 | 17457 | -2.544 | -0.5335 | No | ||
| 90 | LBR | 17563 | -2.660 | -0.5359 | No | ||
| 91 | NUP214 | 17596 | -2.687 | -0.5349 | No | ||
| 92 | PHKB | 17793 | -2.885 | -0.5413 | Yes | ||
| 93 | TADA3L | 17805 | -2.899 | -0.5391 | Yes | ||
| 94 | HDAC5 | 17835 | -2.933 | -0.5378 | Yes | ||
| 95 | PEX1 | 17857 | -2.951 | -0.5360 | Yes | ||
| 96 | TPST2 | 17869 | -2.961 | -0.5338 | Yes | ||
| 97 | RCBTB2 | 17947 | -3.052 | -0.5346 | Yes | ||
| 98 | MAPK3 | 17962 | -3.067 | -0.5324 | Yes | ||
| 99 | FKBP1A | 17973 | -3.076 | -0.5300 | Yes | ||
| 100 | RGS2 | 18003 | -3.105 | -0.5285 | Yes | ||
| 101 | TBCC | 18075 | -3.184 | -0.5288 | Yes | ||
| 102 | PHKA2 | 18186 | -3.318 | -0.5309 | Yes | ||
| 103 | ARID4A | 18192 | -3.324 | -0.5280 | Yes | ||
| 104 | FADD | 18270 | -3.402 | -0.5285 | Yes | ||
| 105 | ACTN1 | 18273 | -3.406 | -0.5254 | Yes | ||
| 106 | PSCD1 | 18407 | -3.543 | -0.5283 | Yes | ||
| 107 | LRRFIP1 | 18437 | -3.583 | -0.5263 | Yes | ||
| 108 | SPAST | 18458 | -3.611 | -0.5239 | Yes | ||
| 109 | ARHGAP1 | 18490 | -3.648 | -0.5220 | Yes | ||
| 110 | HNRPUL1 | 18530 | -3.691 | -0.5204 | Yes | ||
| 111 | PLCB2 | 18630 | -3.789 | -0.5215 | Yes | ||
| 112 | TIAL1 | 18634 | -3.792 | -0.5181 | Yes | ||
| 113 | PPP1R12B | 18636 | -3.794 | -0.5147 | Yes | ||
| 114 | TOB1 | 18651 | -3.809 | -0.5119 | Yes | ||
| 115 | RALBP1 | 18685 | -3.861 | -0.5098 | Yes | ||
| 116 | PHC2 | 18727 | -3.915 | -0.5081 | Yes | ||
| 117 | TSC22D3 | 18787 | -3.985 | -0.5072 | Yes | ||
| 118 | SFRS4 | 18800 | -3.999 | -0.5041 | Yes | ||
| 119 | APAF1 | 18861 | -4.099 | -0.5030 | Yes | ||
| 120 | GNAS | 18973 | -4.235 | -0.5043 | Yes | ||
| 121 | ST8SIA4 | 18988 | -4.256 | -0.5010 | Yes | ||
| 122 | RAF1 | 19074 | -4.383 | -0.5009 | Yes | ||
| 123 | PRMT2 | 19127 | -4.466 | -0.4992 | Yes | ||
| 124 | PRCP | 19132 | -4.471 | -0.4953 | Yes | ||
| 125 | CDIPT | 19182 | -4.552 | -0.4933 | Yes | ||
| 126 | STX16 | 19183 | -4.553 | -0.4891 | Yes | ||
| 127 | WAS | 19208 | -4.589 | -0.4860 | Yes | ||
| 128 | PPFIA1 | 19305 | -4.733 | -0.4861 | Yes | ||
| 129 | PITPNM1 | 19330 | -4.782 | -0.4828 | Yes | ||
| 130 | STAT6 | 19356 | -4.818 | -0.4795 | Yes | ||
| 131 | RARA | 19397 | -4.888 | -0.4769 | Yes | ||
| 132 | ULK1 | 19480 | -5.004 | -0.4761 | Yes | ||
| 133 | SMARCA2 | 19497 | -5.028 | -0.4722 | Yes | ||
| 134 | TAF4 | 19624 | -5.239 | -0.4732 | Yes | ||
| 135 | JAK1 | 19675 | -5.293 | -0.4706 | Yes | ||
| 136 | USP15 | 19714 | -5.353 | -0.4674 | Yes | ||
| 137 | RERE | 19778 | -5.444 | -0.4653 | Yes | ||
| 138 | PECAM1 | 19802 | -5.496 | -0.4614 | Yes | ||
| 139 | IQGAP1 | 19808 | -5.506 | -0.4565 | Yes | ||
| 140 | GTF2B | 19876 | -5.626 | -0.4544 | Yes | ||
| 141 | ACVR1B | 19911 | -5.677 | -0.4508 | Yes | ||
| 142 | PPP1R12A | 19974 | -5.824 | -0.4483 | Yes | ||
| 143 | TUBA1 | 20075 | -6.030 | -0.4474 | Yes | ||
| 144 | KNS2 | 20160 | -6.194 | -0.4455 | Yes | ||
| 145 | GNAQ | 20205 | -6.283 | -0.4418 | Yes | ||
| 146 | PRKAR1A | 20283 | -6.451 | -0.4394 | Yes | ||
| 147 | ITGB2 | 20306 | -6.502 | -0.4345 | Yes | ||
| 148 | RABIF | 20312 | -6.512 | -0.4287 | Yes | ||
| 149 | PTK9L | 20336 | -6.564 | -0.4237 | Yes | ||
| 150 | LTB | 20386 | -6.693 | -0.4198 | Yes | ||
| 151 | MAP3K3 | 20422 | -6.791 | -0.4152 | Yes | ||
| 152 | MYO9B | 20456 | -6.884 | -0.4104 | Yes | ||
| 153 | TBPL1 | 20470 | -6.907 | -0.4047 | Yes | ||
| 154 | OGT | 20476 | -6.916 | -0.3986 | Yes | ||
| 155 | ACTN4 | 20478 | -6.921 | -0.3922 | Yes | ||
| 156 | CAPZB | 20553 | -7.080 | -0.3891 | Yes | ||
| 157 | FOXO1A | 20562 | -7.105 | -0.3830 | Yes | ||
| 158 | LASP1 | 20671 | -7.411 | -0.3812 | Yes | ||
| 159 | ADRBK1 | 20703 | -7.508 | -0.3757 | Yes | ||
| 160 | INPP5D | 20758 | -7.665 | -0.3711 | Yes | ||
| 161 | RASGRP2 | 20770 | -7.709 | -0.3646 | Yes | ||
| 162 | RASA1 | 20833 | -7.927 | -0.3601 | Yes | ||
| 163 | IDS | 20834 | -7.930 | -0.3528 | Yes | ||
| 164 | GDI2 | 20839 | -7.950 | -0.3457 | Yes | ||
| 165 | CUGBP2 | 20871 | -8.070 | -0.3397 | Yes | ||
| 166 | PPM1A | 20886 | -8.120 | -0.3329 | Yes | ||
| 167 | TGFB1 | 20897 | -8.163 | -0.3259 | Yes | ||
| 168 | NCOA1 | 20923 | -8.265 | -0.3195 | Yes | ||
| 169 | PPP1CC | 20962 | -8.421 | -0.3135 | Yes | ||
| 170 | IGF2R | 21017 | -8.646 | -0.3080 | Yes | ||
| 171 | GNAI2 | 21032 | -8.712 | -0.3007 | Yes | ||
| 172 | SLC23A2 | 21042 | -8.735 | -0.2931 | Yes | ||
| 173 | TAOK3 | 21049 | -8.758 | -0.2853 | Yes | ||
| 174 | ABCG1 | 21060 | -8.796 | -0.2777 | Yes | ||
| 175 | NUP153 | 21076 | -8.855 | -0.2702 | Yes | ||
| 176 | FLI1 | 21086 | -8.875 | -0.2625 | Yes | ||
| 177 | ARHGAP4 | 21091 | -8.891 | -0.2545 | Yes | ||
| 178 | SPTLC1 | 21184 | -9.287 | -0.2502 | Yes | ||
| 179 | UBN1 | 21204 | -9.393 | -0.2425 | Yes | ||
| 180 | ARHGDIB | 21340 | -10.120 | -0.2394 | Yes | ||
| 181 | CDC2L5 | 21362 | -10.257 | -0.2309 | Yes | ||
| 182 | RBM5 | 21367 | -10.275 | -0.2217 | Yes | ||
| 183 | DOCK2 | 21381 | -10.369 | -0.2128 | Yes | ||
| 184 | CREBBP | 21411 | -10.605 | -0.2044 | Yes | ||
| 185 | GTF3C1 | 21417 | -10.657 | -0.1948 | Yes | ||
| 186 | GSK3B | 21455 | -10.935 | -0.1865 | Yes | ||
| 187 | MAP2K4 | 21466 | -10.991 | -0.1768 | Yes | ||
| 188 | SRPK2 | 21476 | -11.046 | -0.1671 | Yes | ||
| 189 | EDG6 | 21494 | -11.278 | -0.1575 | Yes | ||
| 190 | MTMR3 | 21499 | -11.309 | -0.1473 | Yes | ||
| 191 | FLOT2 | 21523 | -11.494 | -0.1378 | Yes | ||
| 192 | ADD3 | 21539 | -11.595 | -0.1279 | Yes | ||
| 193 | STK24 | 21610 | -12.437 | -0.1197 | Yes | ||
| 194 | CORO1A | 21631 | -12.679 | -0.1090 | Yes | ||
| 195 | RGS14 | 21660 | -13.135 | -0.0982 | Yes | ||
| 196 | YY1 | 21700 | -13.773 | -0.0873 | Yes | ||
| 197 | STK38 | 21758 | -14.684 | -0.0765 | Yes | ||
| 198 | ARHGEF1 | 21850 | -17.519 | -0.0646 | Yes | ||
| 199 | MARK2 | 21858 | -17.748 | -0.0486 | Yes | ||
| 200 | PRKCB1 | 21866 | -17.985 | -0.0324 | Yes | ||
| 201 | ITPKB | 21894 | -19.628 | -0.0156 | Yes | ||
| 202 | CDKN2D | 21895 | -19.642 | 0.0024 | Yes |
