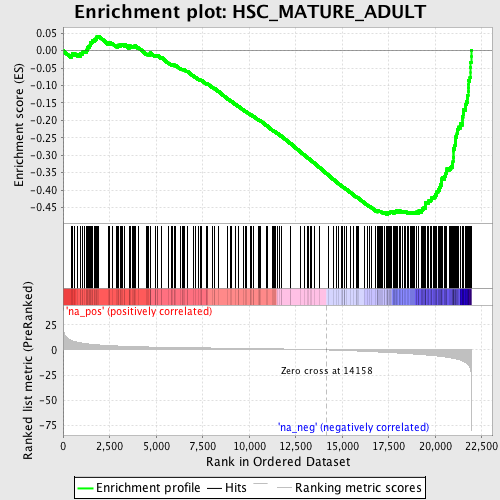
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSC_MATURE_ADULT |
| Enrichment Score (ES) | -0.4699501 |
| Normalized Enrichment Score (NES) | -2.1096773 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0020983738 |
| FWER p-Value | 0.035 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ACSL1 | 433 | 9.490 | -0.0130 | No | ||
| 2 | SUCLG2 | 481 | 9.156 | -0.0084 | No | ||
| 3 | COPS3 | 584 | 8.501 | -0.0068 | No | ||
| 4 | DSTN | 775 | 7.673 | -0.0099 | No | ||
| 5 | TM4SF4 | 919 | 7.146 | -0.0112 | No | ||
| 6 | PIGQ | 936 | 7.078 | -0.0067 | No | ||
| 7 | SH3TC2 | 1040 | 6.769 | -0.0065 | No | ||
| 8 | CTSB | 1065 | 6.681 | -0.0027 | No | ||
| 9 | FGR | 1144 | 6.449 | -0.0015 | No | ||
| 10 | TSPAN17 | 1253 | 6.144 | -0.0019 | No | ||
| 11 | GPX4 | 1265 | 6.125 | 0.0021 | No | ||
| 12 | PEX16 | 1311 | 6.009 | 0.0045 | No | ||
| 13 | TFDP2 | 1321 | 5.981 | 0.0085 | No | ||
| 14 | RNF144 | 1340 | 5.936 | 0.0120 | No | ||
| 15 | ACP1 | 1404 | 5.803 | 0.0134 | No | ||
| 16 | EGFR | 1441 | 5.720 | 0.0160 | No | ||
| 17 | HEMGN | 1470 | 5.640 | 0.0188 | No | ||
| 18 | TMEM16K | 1477 | 5.625 | 0.0227 | No | ||
| 19 | VCL | 1519 | 5.568 | 0.0249 | No | ||
| 20 | ABCB6 | 1580 | 5.452 | 0.0262 | No | ||
| 21 | NAT9 | 1593 | 5.422 | 0.0297 | No | ||
| 22 | LGMN | 1693 | 5.259 | 0.0290 | No | ||
| 23 | LGALS3 | 1704 | 5.240 | 0.0324 | No | ||
| 24 | LAMP2 | 1758 | 5.151 | 0.0337 | No | ||
| 25 | CYBB | 1786 | 5.104 | 0.0363 | No | ||
| 26 | MTHFD2 | 1800 | 5.075 | 0.0394 | No | ||
| 27 | WIPI1 | 1834 | 5.033 | 0.0416 | No | ||
| 28 | KLF11 | 1908 | 4.929 | 0.0419 | No | ||
| 29 | ELA1 | 2459 | 4.255 | 0.0196 | No | ||
| 30 | PTDSS2 | 2465 | 4.243 | 0.0226 | No | ||
| 31 | PRC1 | 2491 | 4.219 | 0.0245 | No | ||
| 32 | ST3GAL5 | 2636 | 4.067 | 0.0209 | No | ||
| 33 | ASNS | 2861 | 3.899 | 0.0134 | No | ||
| 34 | KIAA1715 | 2941 | 3.837 | 0.0126 | No | ||
| 35 | MGST1 | 2970 | 3.821 | 0.0141 | No | ||
| 36 | NUP50 | 2982 | 3.811 | 0.0164 | No | ||
| 37 | LPHN3 | 3067 | 3.754 | 0.0153 | No | ||
| 38 | RAB28 | 3075 | 3.752 | 0.0178 | No | ||
| 39 | ATP8A1 | 3125 | 3.712 | 0.0183 | No | ||
| 40 | LOXL2 | 3208 | 3.648 | 0.0172 | No | ||
| 41 | CCNA2 | 3305 | 3.575 | 0.0154 | No | ||
| 42 | GARNL4 | 3306 | 3.574 | 0.0180 | No | ||
| 43 | CCDC99 | 3545 | 3.409 | 0.0096 | No | ||
| 44 | AHNAK | 3551 | 3.402 | 0.0119 | No | ||
| 45 | TCEA1 | 3554 | 3.397 | 0.0143 | No | ||
| 46 | TIRAP | 3620 | 3.349 | 0.0137 | No | ||
| 47 | NUDT4 | 3703 | 3.308 | 0.0124 | No | ||
| 48 | PLCB1 | 3790 | 3.260 | 0.0108 | No | ||
| 49 | CDK8 | 3816 | 3.244 | 0.0121 | No | ||
| 50 | ADIPOR1 | 3835 | 3.237 | 0.0137 | No | ||
| 51 | TMCC2 | 3864 | 3.222 | 0.0147 | No | ||
| 52 | HSPA12A | 4056 | 3.112 | 0.0082 | No | ||
| 53 | HTATIP2 | 4497 | 2.915 | -0.0099 | No | ||
| 54 | TMPO | 4551 | 2.891 | -0.0102 | No | ||
| 55 | BMPR1A | 4580 | 2.883 | -0.0094 | No | ||
| 56 | SLC11A1 | 4672 | 2.846 | -0.0115 | No | ||
| 57 | CD14 | 4674 | 2.846 | -0.0095 | No | ||
| 58 | ORM2 | 4683 | 2.842 | -0.0077 | No | ||
| 59 | HRB | 4689 | 2.840 | -0.0059 | No | ||
| 60 | MARCH8 | 4938 | 2.741 | -0.0153 | No | ||
| 61 | BMX | 4945 | 2.739 | -0.0135 | No | ||
| 62 | SLC4A1 | 4967 | 2.733 | -0.0125 | No | ||
| 63 | C3 | 5052 | 2.705 | -0.0144 | No | ||
| 64 | CDKN3 | 5062 | 2.699 | -0.0128 | No | ||
| 65 | UPB1 | 5286 | 2.627 | -0.0211 | No | ||
| 66 | ARL2BP | 5310 | 2.621 | -0.0203 | No | ||
| 67 | GOLPH4 | 5675 | 2.502 | -0.0352 | No | ||
| 68 | SLC16A1 | 5824 | 2.456 | -0.0402 | No | ||
| 69 | PBX1 | 5844 | 2.448 | -0.0393 | No | ||
| 70 | EIF5A2 | 5875 | 2.438 | -0.0389 | No | ||
| 71 | ACTA2 | 5985 | 2.401 | -0.0421 | No | ||
| 72 | SQRDL | 5995 | 2.398 | -0.0408 | No | ||
| 73 | TNFSF13B | 6062 | 2.377 | -0.0421 | No | ||
| 74 | MOSPD1 | 6281 | 2.309 | -0.0504 | No | ||
| 75 | PILRA | 6389 | 2.283 | -0.0537 | No | ||
| 76 | ADAM10 | 6405 | 2.277 | -0.0527 | No | ||
| 77 | S100A6 | 6457 | 2.262 | -0.0534 | No | ||
| 78 | MRPL53 | 6516 | 2.247 | -0.0544 | No | ||
| 79 | TSPAN8 | 6661 | 2.205 | -0.0594 | No | ||
| 80 | USP46 | 6692 | 2.198 | -0.0592 | No | ||
| 81 | PPBP | 6984 | 2.123 | -0.0710 | No | ||
| 82 | ARFGEF1 | 7130 | 2.083 | -0.0762 | No | ||
| 83 | AOAH | 7272 | 2.044 | -0.0812 | No | ||
| 84 | KEL | 7362 | 2.021 | -0.0838 | No | ||
| 85 | SLC40A1 | 7424 | 2.006 | -0.0851 | No | ||
| 86 | IGSF6 | 7438 | 2.002 | -0.0843 | No | ||
| 87 | GCNT1 | 7696 | 1.931 | -0.0947 | No | ||
| 88 | DMXL2 | 7719 | 1.925 | -0.0943 | No | ||
| 89 | ARHGAP19 | 7774 | 1.912 | -0.0954 | No | ||
| 90 | STX17 | 8003 | 1.857 | -0.1045 | No | ||
| 91 | PVALB | 8117 | 1.833 | -0.1084 | No | ||
| 92 | EMR4 | 8151 | 1.826 | -0.1086 | No | ||
| 93 | UGP2 | 8358 | 1.778 | -0.1168 | No | ||
| 94 | SPECC1 | 8853 | 1.660 | -0.1383 | No | ||
| 95 | EMR1 | 8968 | 1.632 | -0.1424 | No | ||
| 96 | ENDOD1 | 9045 | 1.615 | -0.1447 | No | ||
| 97 | CTSH | 9281 | 1.556 | -0.1544 | No | ||
| 98 | TLR6 | 9407 | 1.528 | -0.1590 | No | ||
| 99 | TRIB1 | 9436 | 1.520 | -0.1592 | No | ||
| 100 | CDR2 | 9667 | 1.470 | -0.1688 | No | ||
| 101 | RAD51C | 9814 | 1.430 | -0.1744 | No | ||
| 102 | CLEC4E | 9837 | 1.426 | -0.1744 | No | ||
| 103 | FGD4 | 10058 | 1.368 | -0.1835 | No | ||
| 104 | PROM1 | 10079 | 1.363 | -0.1835 | No | ||
| 105 | ADAM19 | 10140 | 1.349 | -0.1852 | No | ||
| 106 | MLSTD1 | 10248 | 1.323 | -0.1892 | No | ||
| 107 | C1S | 10254 | 1.322 | -0.1884 | No | ||
| 108 | HAGH | 10476 | 1.268 | -0.1977 | No | ||
| 109 | GCA | 10542 | 1.250 | -0.1998 | No | ||
| 110 | MRC1 | 10555 | 1.247 | -0.1994 | No | ||
| 111 | SLCO4C1 | 10593 | 1.238 | -0.2002 | No | ||
| 112 | SGOL2 | 10906 | 1.158 | -0.2138 | No | ||
| 113 | BNIP3L | 10963 | 1.146 | -0.2155 | No | ||
| 114 | TMEM56 | 11262 | 1.066 | -0.2285 | No | ||
| 115 | ITGAM | 11326 | 1.048 | -0.2306 | No | ||
| 116 | PLA2G2F | 11380 | 1.029 | -0.2323 | No | ||
| 117 | PPP1R3D | 11431 | 1.013 | -0.2339 | No | ||
| 118 | RSAD2 | 11531 | 0.984 | -0.2377 | No | ||
| 119 | XLKD1 | 11641 | 0.954 | -0.2420 | No | ||
| 120 | HEBP1 | 11716 | 0.930 | -0.2448 | No | ||
| 121 | CD9 | 11727 | 0.926 | -0.2445 | No | ||
| 122 | CES1 | 12223 | 0.774 | -0.2668 | No | ||
| 123 | RAB3D | 12236 | 0.771 | -0.2668 | No | ||
| 124 | CCRN4L | 12773 | 0.597 | -0.2911 | No | ||
| 125 | BMP2K | 12987 | 0.517 | -0.3006 | No | ||
| 126 | ANKRD9 | 13133 | 0.463 | -0.3069 | No | ||
| 127 | RHD | 13175 | 0.446 | -0.3085 | No | ||
| 128 | GTPBP6 | 13304 | 0.393 | -0.3141 | No | ||
| 129 | STAB2 | 13351 | 0.375 | -0.3159 | No | ||
| 130 | ANKRD43 | 13509 | 0.313 | -0.3230 | No | ||
| 131 | LRG1 | 13767 | 0.186 | -0.3347 | No | ||
| 132 | P2RY13 | 14262 | -0.070 | -0.3574 | No | ||
| 133 | GYPA | 14532 | -0.236 | -0.3697 | No | ||
| 134 | PQLC3 | 14689 | -0.333 | -0.3766 | No | ||
| 135 | HOOK3 | 14808 | -0.422 | -0.3818 | No | ||
| 136 | WDR32 | 14933 | -0.494 | -0.3871 | No | ||
| 137 | SLC7A8 | 15005 | -0.539 | -0.3900 | No | ||
| 138 | MMP8 | 15020 | -0.547 | -0.3903 | No | ||
| 139 | DEDD2 | 15109 | -0.619 | -0.3939 | No | ||
| 140 | PIK3CB | 15122 | -0.630 | -0.3940 | No | ||
| 141 | DEGS1 | 15124 | -0.631 | -0.3935 | No | ||
| 142 | GPATC1 | 15243 | -0.705 | -0.3985 | No | ||
| 143 | GRINA | 15453 | -0.848 | -0.4075 | No | ||
| 144 | UBE2L6 | 15616 | -0.953 | -0.4143 | No | ||
| 145 | SLC25A37 | 15757 | -1.054 | -0.4199 | No | ||
| 146 | RNF19 | 15767 | -1.058 | -0.4196 | No | ||
| 147 | CD226 | 15791 | -1.073 | -0.4198 | No | ||
| 148 | SLC30A10 | 15894 | -1.148 | -0.4237 | No | ||
| 149 | CLIC6 | 16193 | -1.404 | -0.4364 | No | ||
| 150 | LY75 | 16366 | -1.548 | -0.4432 | No | ||
| 151 | SCRN3 | 16474 | -1.636 | -0.4470 | No | ||
| 152 | JMJD2C | 16572 | -1.727 | -0.4502 | No | ||
| 153 | MLSTD2 | 16793 | -1.923 | -0.4589 | No | ||
| 154 | DSU | 16875 | -2.009 | -0.4611 | No | ||
| 155 | ANXA2 | 16913 | -2.039 | -0.4613 | No | ||
| 156 | LRRC28 | 16916 | -2.041 | -0.4599 | No | ||
| 157 | SNCA | 16938 | -2.060 | -0.4594 | No | ||
| 158 | NR3C1 | 17008 | -2.137 | -0.4610 | No | ||
| 159 | SORT1 | 17032 | -2.158 | -0.4604 | No | ||
| 160 | FMO5 | 17121 | -2.238 | -0.4629 | No | ||
| 161 | SLK | 17186 | -2.292 | -0.4641 | No | ||
| 162 | RBMS1 | 17267 | -2.356 | -0.4661 | No | ||
| 163 | TXNDC1 | 17285 | -2.372 | -0.4651 | No | ||
| 164 | XPO7 | 17391 | -2.472 | -0.4681 | Yes | ||
| 165 | MPP1 | 17422 | -2.508 | -0.4677 | Yes | ||
| 166 | SGK | 17430 | -2.515 | -0.4661 | Yes | ||
| 167 | SYAP1 | 17444 | -2.532 | -0.4648 | Yes | ||
| 168 | CAB39 | 17508 | -2.607 | -0.4658 | Yes | ||
| 169 | UBLCP1 | 17523 | -2.624 | -0.4645 | Yes | ||
| 170 | LBR | 17563 | -2.660 | -0.4644 | Yes | ||
| 171 | HIATL1 | 17589 | -2.682 | -0.4635 | Yes | ||
| 172 | RFFL | 17612 | -2.699 | -0.4626 | Yes | ||
| 173 | CPD | 17633 | -2.723 | -0.4615 | Yes | ||
| 174 | PRPF18 | 17741 | -2.823 | -0.4643 | Yes | ||
| 175 | MYO9A | 17748 | -2.831 | -0.4625 | Yes | ||
| 176 | SLC1A5 | 17823 | -2.923 | -0.4638 | Yes | ||
| 177 | ASB7 | 17825 | -2.925 | -0.4616 | Yes | ||
| 178 | SOAT1 | 17864 | -2.959 | -0.4612 | Yes | ||
| 179 | NCAM1 | 17916 | -3.014 | -0.4613 | Yes | ||
| 180 | ARPC5 | 17926 | -3.022 | -0.4595 | Yes | ||
| 181 | TRIM59 | 17986 | -3.091 | -0.4600 | Yes | ||
| 182 | TMEM9B | 18073 | -3.182 | -0.4616 | Yes | ||
| 183 | UBADC1 | 18080 | -3.193 | -0.4595 | Yes | ||
| 184 | ARFIP1 | 18139 | -3.258 | -0.4598 | Yes | ||
| 185 | PPP3R1 | 18216 | -3.345 | -0.4608 | Yes | ||
| 186 | RAB24 | 18259 | -3.390 | -0.4602 | Yes | ||
| 187 | MGST3 | 18358 | -3.487 | -0.4622 | Yes | ||
| 188 | SACM1L | 18392 | -3.525 | -0.4611 | Yes | ||
| 189 | IFIT1 | 18521 | -3.678 | -0.4643 | Yes | ||
| 190 | RNF185 | 18578 | -3.732 | -0.4641 | Yes | ||
| 191 | SDCBP | 18656 | -3.818 | -0.4649 | Yes | ||
| 192 | RABAC1 | 18694 | -3.870 | -0.4637 | Yes | ||
| 193 | RB1 | 18763 | -3.958 | -0.4639 | Yes | ||
| 194 | RIOK3 | 18844 | -4.070 | -0.4646 | Yes | ||
| 195 | BRMS1 | 18885 | -4.131 | -0.4634 | Yes | ||
| 196 | EIF2AK3 | 18978 | -4.241 | -0.4645 | Yes | ||
| 197 | SNX13 | 19009 | -4.292 | -0.4627 | Yes | ||
| 198 | INPP5A | 19100 | -4.418 | -0.4636 | Yes | ||
| 199 | CAST | 19103 | -4.424 | -0.4604 | Yes | ||
| 200 | MON1A | 19120 | -4.458 | -0.4579 | Yes | ||
| 201 | PTEN | 19245 | -4.642 | -0.4602 | Yes | ||
| 202 | HACE1 | 19267 | -4.675 | -0.4577 | Yes | ||
| 203 | OTUD5 | 19306 | -4.735 | -0.4559 | Yes | ||
| 204 | PI4K2B | 19319 | -4.764 | -0.4530 | Yes | ||
| 205 | MAP1LC3B | 19359 | -4.824 | -0.4512 | Yes | ||
| 206 | MTF1 | 19396 | -4.885 | -0.4492 | Yes | ||
| 207 | EPS15 | 19457 | -4.980 | -0.4483 | Yes | ||
| 208 | CPEB4 | 19462 | -4.985 | -0.4448 | Yes | ||
| 209 | FOXJ2 | 19463 | -4.985 | -0.4412 | Yes | ||
| 210 | SLC7A11 | 19476 | -5.002 | -0.4380 | Yes | ||
| 211 | LNPEP | 19484 | -5.011 | -0.4346 | Yes | ||
| 212 | SERINC5 | 19584 | -5.165 | -0.4354 | Yes | ||
| 213 | RNF141 | 19641 | -5.256 | -0.4341 | Yes | ||
| 214 | HSPA4L | 19656 | -5.273 | -0.4308 | Yes | ||
| 215 | WDR26 | 19719 | -5.360 | -0.4297 | Yes | ||
| 216 | USP25 | 19767 | -5.434 | -0.4279 | Yes | ||
| 217 | KLHL2 | 19784 | -5.468 | -0.4246 | Yes | ||
| 218 | CSPP1 | 19814 | -5.518 | -0.4218 | Yes | ||
| 219 | FBXL5 | 19883 | -5.637 | -0.4208 | Yes | ||
| 220 | STX2 | 19933 | -5.735 | -0.4188 | Yes | ||
| 221 | ANKRD12 | 19987 | -5.842 | -0.4170 | Yes | ||
| 222 | SCARB2 | 20003 | -5.881 | -0.4133 | Yes | ||
| 223 | KIF1B | 20048 | -5.972 | -0.4109 | Yes | ||
| 224 | SERTAD2 | 20051 | -5.975 | -0.4066 | Yes | ||
| 225 | ZFYVE27 | 20085 | -6.047 | -0.4037 | Yes | ||
| 226 | GTPBP2 | 20144 | -6.157 | -0.4018 | Yes | ||
| 227 | SMOX | 20164 | -6.207 | -0.3981 | Yes | ||
| 228 | IFT80 | 20238 | -6.366 | -0.3967 | Yes | ||
| 229 | CEBPG | 20240 | -6.372 | -0.3921 | Yes | ||
| 230 | FN3K | 20265 | -6.422 | -0.3884 | Yes | ||
| 231 | ST6GAL1 | 20276 | -6.437 | -0.3841 | Yes | ||
| 232 | HECTD1 | 20316 | -6.528 | -0.3811 | Yes | ||
| 233 | ATP2C1 | 20325 | -6.547 | -0.3766 | Yes | ||
| 234 | USP32 | 20341 | -6.574 | -0.3725 | Yes | ||
| 235 | GAPVD1 | 20350 | -6.600 | -0.3680 | Yes | ||
| 236 | CMAS | 20379 | -6.663 | -0.3643 | Yes | ||
| 237 | PGLYRP1 | 20485 | -6.937 | -0.3641 | Yes | ||
| 238 | KCTD2 | 20500 | -6.961 | -0.3596 | Yes | ||
| 239 | TRAK2 | 20547 | -7.070 | -0.3564 | Yes | ||
| 240 | WDR40A | 20564 | -7.109 | -0.3519 | Yes | ||
| 241 | UBE4A | 20592 | -7.185 | -0.3479 | Yes | ||
| 242 | ARL6IP1 | 20620 | -7.277 | -0.3437 | Yes | ||
| 243 | C1GALT1 | 20624 | -7.283 | -0.3385 | Yes | ||
| 244 | GRIPAP1 | 20741 | -7.593 | -0.3382 | Yes | ||
| 245 | LAMP1 | 20819 | -7.881 | -0.3360 | Yes | ||
| 246 | PQLC1 | 20845 | -7.967 | -0.3312 | Yes | ||
| 247 | MARCH2 | 20916 | -8.222 | -0.3284 | Yes | ||
| 248 | AP2A2 | 20919 | -8.237 | -0.3224 | Yes | ||
| 249 | MXI1 | 20947 | -8.353 | -0.3175 | Yes | ||
| 250 | ALAS2 | 20954 | -8.381 | -0.3115 | Yes | ||
| 251 | TNRC6B | 20955 | -8.389 | -0.3053 | Yes | ||
| 252 | ETS1 | 20973 | -8.470 | -0.2999 | Yes | ||
| 253 | DLEU2 | 20974 | -8.472 | -0.2936 | Yes | ||
| 254 | MTSS1 | 20975 | -8.479 | -0.2873 | Yes | ||
| 255 | ST3GAL6 | 20987 | -8.532 | -0.2815 | Yes | ||
| 256 | POLG | 21020 | -8.661 | -0.2766 | Yes | ||
| 257 | MXD1 | 21051 | -8.766 | -0.2715 | Yes | ||
| 258 | ABLIM1 | 21068 | -8.821 | -0.2657 | Yes | ||
| 259 | GDPD3 | 21075 | -8.851 | -0.2595 | Yes | ||
| 260 | PDPN | 21096 | -8.916 | -0.2538 | Yes | ||
| 261 | HERC4 | 21101 | -8.924 | -0.2474 | Yes | ||
| 262 | YPEL5 | 21147 | -9.131 | -0.2427 | Yes | ||
| 263 | SAMD9L | 21163 | -9.197 | -0.2366 | Yes | ||
| 264 | LMTK2 | 21182 | -9.262 | -0.2306 | Yes | ||
| 265 | ZSWIM6 | 21205 | -9.397 | -0.2247 | Yes | ||
| 266 | CDC42EP3 | 21234 | -9.551 | -0.2189 | Yes | ||
| 267 | PICALM | 21330 | -10.065 | -0.2158 | Yes | ||
| 268 | LIMS1 | 21355 | -10.221 | -0.2094 | Yes | ||
| 269 | MAP4K5 | 21442 | -10.831 | -0.2054 | Yes | ||
| 270 | SYNJ1 | 21447 | -10.859 | -0.1975 | Yes | ||
| 271 | RNF167 | 21454 | -10.933 | -0.1897 | Yes | ||
| 272 | MKRN1 | 21488 | -11.213 | -0.1829 | Yes | ||
| 273 | BAZ2B | 21508 | -11.364 | -0.1754 | Yes | ||
| 274 | GSPT1 | 21530 | -11.532 | -0.1679 | Yes | ||
| 275 | USP7 | 21623 | -12.586 | -0.1628 | Yes | ||
| 276 | HERPUD2 | 21637 | -12.814 | -0.1539 | Yes | ||
| 277 | SEMA4A | 21668 | -13.174 | -0.1456 | Yes | ||
| 278 | E2F2 | 21730 | -14.272 | -0.1378 | Yes | ||
| 279 | TCP11L2 | 21741 | -14.504 | -0.1276 | Yes | ||
| 280 | STK38 | 21758 | -14.684 | -0.1175 | Yes | ||
| 281 | ADD1 | 21766 | -14.850 | -0.1068 | Yes | ||
| 282 | WHSC1L1 | 21772 | -15.077 | -0.0959 | Yes | ||
| 283 | KLF3 | 21779 | -15.184 | -0.0850 | Yes | ||
| 284 | STK17B | 21860 | -17.835 | -0.0755 | Yes | ||
| 285 | LRRC35 | 21893 | -19.607 | -0.0624 | Yes | ||
| 286 | CDKN2D | 21895 | -19.642 | -0.0480 | Yes | ||
| 287 | KLF7 | 21904 | -20.512 | -0.0332 | Yes | ||
| 288 | CCND3 | 21924 | -22.833 | -0.0172 | Yes | ||
| 289 | DCK | 21929 | -24.615 | 0.0008 | Yes |
