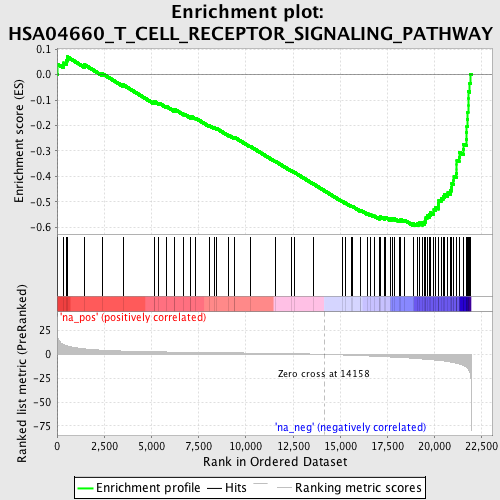
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.5949742 |
| Normalized Enrichment Score (NES) | -2.2174888 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.5248005E-4 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL5 | 11 | 22.605 | 0.0418 | No | ||
| 2 | CDK4 | 343 | 10.371 | 0.0461 | No | ||
| 3 | HRAS | 510 | 8.966 | 0.0553 | No | ||
| 4 | NCK2 | 543 | 8.728 | 0.0702 | No | ||
| 5 | PIK3R2 | 1448 | 5.700 | 0.0395 | No | ||
| 6 | NFATC2 | 2387 | 4.322 | 0.0047 | No | ||
| 7 | KRAS | 3504 | 3.433 | -0.0399 | No | ||
| 8 | PPP3R2 | 5146 | 2.673 | -0.1100 | No | ||
| 9 | CBLC | 5155 | 2.667 | -0.1053 | No | ||
| 10 | NFATC4 | 5397 | 2.593 | -0.1115 | No | ||
| 11 | SOS1 | 5789 | 2.466 | -0.1248 | No | ||
| 12 | PIK3R1 | 6230 | 2.327 | -0.1406 | No | ||
| 13 | IFNG | 6243 | 2.320 | -0.1368 | No | ||
| 14 | PAK3 | 6721 | 2.192 | -0.1545 | No | ||
| 15 | IL10 | 7078 | 2.096 | -0.1668 | No | ||
| 16 | NFKB1 | 7091 | 2.093 | -0.1635 | No | ||
| 17 | CDC42 | 7329 | 2.030 | -0.1705 | No | ||
| 18 | PIK3R3 | 8058 | 1.844 | -0.2004 | No | ||
| 19 | VAV3 | 8314 | 1.784 | -0.2087 | No | ||
| 20 | PAK7 | 8468 | 1.750 | -0.2124 | No | ||
| 21 | PPP3CA | 9099 | 1.601 | -0.2382 | No | ||
| 22 | PPP3CC | 9402 | 1.530 | -0.2492 | No | ||
| 23 | NFKBIB | 9422 | 1.525 | -0.2472 | No | ||
| 24 | NFKBIE | 10229 | 1.326 | -0.2816 | No | ||
| 25 | PDCD1 | 11546 | 0.981 | -0.3400 | No | ||
| 26 | IL4 | 12402 | 0.724 | -0.3777 | No | ||
| 27 | JUN | 12565 | 0.666 | -0.3839 | No | ||
| 28 | CSF2 | 13564 | 0.288 | -0.4290 | No | ||
| 29 | PIK3CB | 15122 | -0.630 | -0.4991 | No | ||
| 30 | CD40LG | 15266 | -0.717 | -0.5043 | No | ||
| 31 | GRB2 | 15610 | -0.952 | -0.5182 | No | ||
| 32 | LAT | 15646 | -0.971 | -0.5179 | No | ||
| 33 | TNF | 16079 | -1.307 | -0.5353 | No | ||
| 34 | PAK4 | 16081 | -1.308 | -0.5329 | No | ||
| 35 | ZAP70 | 16444 | -1.608 | -0.5464 | No | ||
| 36 | SOS2 | 16589 | -1.746 | -0.5497 | No | ||
| 37 | PAK6 | 16798 | -1.927 | -0.5556 | No | ||
| 38 | AKT1 | 17066 | -2.193 | -0.5637 | No | ||
| 39 | RHOA | 17075 | -2.199 | -0.5600 | No | ||
| 40 | NCK1 | 17148 | -2.262 | -0.5590 | No | ||
| 41 | PTPN6 | 17353 | -2.437 | -0.5638 | No | ||
| 42 | MAP3K14 | 17377 | -2.462 | -0.5602 | No | ||
| 43 | CHUK | 17643 | -2.730 | -0.5673 | No | ||
| 44 | PAK1 | 17746 | -2.826 | -0.5666 | No | ||
| 45 | MAP3K8 | 17860 | -2.954 | -0.5663 | No | ||
| 46 | AKT3 | 18133 | -3.248 | -0.5726 | No | ||
| 47 | PPP3R1 | 18216 | -3.345 | -0.5701 | No | ||
| 48 | IKBKG | 18397 | -3.530 | -0.5717 | No | ||
| 49 | PAK2 | 18865 | -4.102 | -0.5854 | No | ||
| 50 | CTLA4 | 19075 | -4.385 | -0.5868 | Yes | ||
| 51 | FOS | 19178 | -4.546 | -0.5829 | Yes | ||
| 52 | CD3G | 19364 | -4.837 | -0.5823 | Yes | ||
| 53 | NFKB2 | 19492 | -5.020 | -0.5787 | Yes | ||
| 54 | PTPRC | 19517 | -5.062 | -0.5703 | Yes | ||
| 55 | NFAT5 | 19545 | -5.113 | -0.5620 | Yes | ||
| 56 | CBLB | 19603 | -5.196 | -0.5549 | Yes | ||
| 57 | IKBKB | 19718 | -5.359 | -0.5500 | Yes | ||
| 58 | PDK1 | 19800 | -5.491 | -0.5435 | Yes | ||
| 59 | GRAP2 | 19967 | -5.818 | -0.5402 | Yes | ||
| 60 | CD3D | 19969 | -5.819 | -0.5293 | Yes | ||
| 61 | VAV1 | 20066 | -6.018 | -0.5224 | Yes | ||
| 62 | PLCG1 | 20195 | -6.259 | -0.5165 | Yes | ||
| 63 | AKT2 | 20209 | -6.297 | -0.5053 | Yes | ||
| 64 | BCL10 | 20231 | -6.352 | -0.4944 | Yes | ||
| 65 | PPP3CB | 20348 | -6.597 | -0.4873 | Yes | ||
| 66 | VAV2 | 20452 | -6.862 | -0.4792 | Yes | ||
| 67 | PIK3CA | 20532 | -7.036 | -0.4696 | Yes | ||
| 68 | ICOS | 20711 | -7.531 | -0.4637 | Yes | ||
| 69 | CD8B | 20855 | -7.998 | -0.4552 | Yes | ||
| 70 | PIK3CD | 20906 | -8.191 | -0.4422 | Yes | ||
| 71 | PRKCQ | 20917 | -8.226 | -0.4272 | Yes | ||
| 72 | PIK3CG | 21024 | -8.675 | -0.4158 | Yes | ||
| 73 | PIK3R5 | 21030 | -8.703 | -0.3997 | Yes | ||
| 74 | NRAS | 21157 | -9.167 | -0.3883 | Yes | ||
| 75 | CD8A | 21185 | -9.290 | -0.3722 | Yes | ||
| 76 | CARD11 | 21186 | -9.295 | -0.3547 | Yes | ||
| 77 | CD4 | 21188 | -9.300 | -0.3374 | Yes | ||
| 78 | FYN | 21301 | -9.911 | -0.3239 | Yes | ||
| 79 | ITK | 21334 | -10.083 | -0.3065 | Yes | ||
| 80 | NFATC1 | 21534 | -11.557 | -0.2940 | Yes | ||
| 81 | CD3E | 21545 | -11.647 | -0.2726 | Yes | ||
| 82 | TEC | 21697 | -13.757 | -0.2537 | Yes | ||
| 83 | CBL | 21707 | -13.898 | -0.2281 | Yes | ||
| 84 | RASGRP1 | 21715 | -14.043 | -0.2021 | Yes | ||
| 85 | NFATC3 | 21743 | -14.538 | -0.1761 | Yes | ||
| 86 | LCP2 | 21764 | -14.815 | -0.1493 | Yes | ||
| 87 | NFKBIA | 21775 | -15.114 | -0.1214 | Yes | ||
| 88 | LCK | 21778 | -15.166 | -0.0931 | Yes | ||
| 89 | CD247 | 21783 | -15.293 | -0.0646 | Yes | ||
| 90 | MALT1 | 21871 | -18.066 | -0.0348 | Yes | ||
| 91 | CD28 | 21902 | -20.380 | 0.0021 | Yes |
