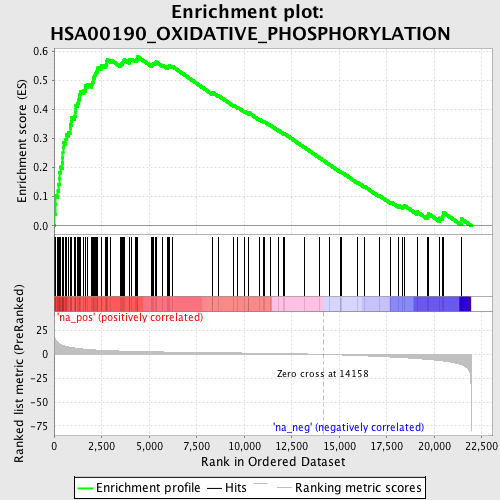
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA00190_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.58144724 |
| Normalized Enrichment Score (NES) | 2.5279484 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP5G1 | 16 | 21.522 | 0.0418 | Yes | ||
| 2 | PPA1 | 53 | 16.446 | 0.0727 | Yes | ||
| 3 | COX5A | 62 | 16.106 | 0.1042 | Yes | ||
| 4 | NDUFA10 | 203 | 11.910 | 0.1213 | Yes | ||
| 5 | UQCRFS1 | 247 | 11.316 | 0.1417 | Yes | ||
| 6 | COX5B | 277 | 11.008 | 0.1621 | Yes | ||
| 7 | NDUFA8 | 288 | 10.858 | 0.1831 | Yes | ||
| 8 | COX8A | 324 | 10.539 | 0.2024 | Yes | ||
| 9 | NDUFV1 | 429 | 9.497 | 0.2164 | Yes | ||
| 10 | UQCRQ | 460 | 9.281 | 0.2334 | Yes | ||
| 11 | NDUFAB1 | 465 | 9.257 | 0.2515 | Yes | ||
| 12 | NDUFA12 | 469 | 9.223 | 0.2696 | Yes | ||
| 13 | SDHD | 480 | 9.166 | 0.2872 | Yes | ||
| 14 | SDHB | 619 | 8.329 | 0.2974 | Yes | ||
| 15 | ATP5L | 634 | 8.280 | 0.3131 | Yes | ||
| 16 | CYC1 | 767 | 7.693 | 0.3223 | Yes | ||
| 17 | NDUFV2 | 840 | 7.433 | 0.3337 | Yes | ||
| 18 | NDUFA11 | 848 | 7.395 | 0.3480 | Yes | ||
| 19 | NDUFB8 | 898 | 7.209 | 0.3600 | Yes | ||
| 20 | UQCR | 932 | 7.096 | 0.3725 | Yes | ||
| 21 | ATP5J | 1098 | 6.550 | 0.3779 | Yes | ||
| 22 | NDUFB9 | 1115 | 6.513 | 0.3900 | Yes | ||
| 23 | NDUFB10 | 1141 | 6.456 | 0.4016 | Yes | ||
| 24 | NDUFS8 | 1146 | 6.442 | 0.4142 | Yes | ||
| 25 | NDUFA9 | 1233 | 6.204 | 0.4225 | Yes | ||
| 26 | NDUFB4 | 1270 | 6.115 | 0.4330 | Yes | ||
| 27 | UQCRC1 | 1327 | 5.967 | 0.4422 | Yes | ||
| 28 | NDUFS3 | 1351 | 5.899 | 0.4528 | Yes | ||
| 29 | COX4I2 | 1409 | 5.797 | 0.4617 | Yes | ||
| 30 | NDUFS6 | 1564 | 5.471 | 0.4654 | Yes | ||
| 31 | ATP5F1 | 1653 | 5.316 | 0.4719 | Yes | ||
| 32 | NDUFB5 | 1667 | 5.294 | 0.4818 | Yes | ||
| 33 | NDUFA7 | 1774 | 5.121 | 0.4871 | Yes | ||
| 34 | ATP5I | 1984 | 4.827 | 0.4870 | Yes | ||
| 35 | COX7C | 2022 | 4.780 | 0.4948 | Yes | ||
| 36 | NDUFC1 | 2054 | 4.741 | 0.5027 | Yes | ||
| 37 | ATP5A1 | 2058 | 4.738 | 0.5120 | Yes | ||
| 38 | TCIRG1 | 2138 | 4.625 | 0.5175 | Yes | ||
| 39 | COX6B1 | 2157 | 4.594 | 0.5257 | Yes | ||
| 40 | ATP5O | 2209 | 4.538 | 0.5324 | Yes | ||
| 41 | NDUFA5 | 2293 | 4.426 | 0.5373 | Yes | ||
| 42 | ATP5E | 2306 | 4.413 | 0.5455 | Yes | ||
| 43 | PPA2 | 2468 | 4.241 | 0.5465 | Yes | ||
| 44 | NDUFB7 | 2510 | 4.199 | 0.5529 | Yes | ||
| 45 | ATP6V0D2 | 2689 | 4.027 | 0.5528 | Yes | ||
| 46 | ATP5J2 | 2754 | 3.980 | 0.5577 | Yes | ||
| 47 | ATP5G3 | 2767 | 3.974 | 0.5650 | Yes | ||
| 48 | ATP5D | 2793 | 3.957 | 0.5717 | Yes | ||
| 49 | ATP6V1G1 | 2991 | 3.806 | 0.5702 | Yes | ||
| 50 | NDUFB6 | 3505 | 3.432 | 0.5535 | Yes | ||
| 51 | UQCRC2 | 3557 | 3.395 | 0.5579 | Yes | ||
| 52 | ATP5G2 | 3603 | 3.359 | 0.5624 | Yes | ||
| 53 | COX6C | 3638 | 3.339 | 0.5675 | Yes | ||
| 54 | NDUFB2 | 3695 | 3.313 | 0.5715 | Yes | ||
| 55 | ATP5C1 | 3949 | 3.174 | 0.5662 | Yes | ||
| 56 | ATP5B | 3951 | 3.174 | 0.5724 | Yes | ||
| 57 | ATP12A | 4055 | 3.112 | 0.5738 | Yes | ||
| 58 | UQCRB | 4258 | 3.014 | 0.5705 | Yes | ||
| 59 | ATP6V0A1 | 4347 | 2.973 | 0.5724 | Yes | ||
| 60 | COX6A2 | 4396 | 2.954 | 0.5760 | Yes | ||
| 61 | ATP4B | 4406 | 2.950 | 0.5814 | Yes | ||
| 62 | COX15 | 5112 | 2.683 | 0.5545 | No | ||
| 63 | NDUFS7 | 5194 | 2.655 | 0.5560 | No | ||
| 64 | NDUFA1 | 5233 | 2.644 | 0.5595 | No | ||
| 65 | ATP6V1G2 | 5313 | 2.620 | 0.5611 | No | ||
| 66 | COX8C | 5364 | 2.603 | 0.5639 | No | ||
| 67 | NDUFA6 | 5703 | 2.494 | 0.5534 | No | ||
| 68 | ATP6V1A | 5970 | 2.405 | 0.5460 | No | ||
| 69 | SDHC | 6017 | 2.390 | 0.5486 | No | ||
| 70 | ATP5H | 6060 | 2.378 | 0.5513 | No | ||
| 71 | ATP4A | 6211 | 2.332 | 0.5491 | No | ||
| 72 | NDUFS2 | 8317 | 1.784 | 0.4562 | No | ||
| 73 | NDUFS5 | 8356 | 1.779 | 0.4580 | No | ||
| 74 | UQCRH | 8656 | 1.702 | 0.4477 | No | ||
| 75 | ATP6V1C1 | 9441 | 1.519 | 0.4148 | No | ||
| 76 | ATP6V1E2 | 9665 | 1.470 | 0.4075 | No | ||
| 77 | ATP6V0B | 10031 | 1.377 | 0.3935 | No | ||
| 78 | COX17 | 10228 | 1.326 | 0.3872 | No | ||
| 79 | COX7A2 | 10247 | 1.323 | 0.3890 | No | ||
| 80 | COX6A1 | 10834 | 1.179 | 0.3645 | No | ||
| 81 | ATP6V0A4 | 11016 | 1.130 | 0.3584 | No | ||
| 82 | NDUFC2 | 11046 | 1.122 | 0.3593 | No | ||
| 83 | NDUFB3 | 11383 | 1.028 | 0.3459 | No | ||
| 84 | NDUFS1 | 11834 | 0.895 | 0.3271 | No | ||
| 85 | COX4I1 | 12097 | 0.812 | 0.3167 | No | ||
| 86 | ATP6V1C2 | 12130 | 0.803 | 0.3168 | No | ||
| 87 | NDUFA4 | 13200 | 0.434 | 0.2688 | No | ||
| 88 | ATP6V1B1 | 13957 | 0.102 | 0.2343 | No | ||
| 89 | COX6B2 | 14493 | -0.217 | 0.2103 | No | ||
| 90 | COX7A1 | 15067 | -0.585 | 0.1852 | No | ||
| 91 | NDUFA2 | 15080 | -0.594 | 0.1858 | No | ||
| 92 | ATP6V1F | 15137 | -0.640 | 0.1845 | No | ||
| 93 | NDUFB11 | 15981 | -1.222 | 0.1483 | No | ||
| 94 | SDHA | 16338 | -1.516 | 0.1350 | No | ||
| 95 | NDUFA13 | 17104 | -2.224 | 0.1044 | No | ||
| 96 | COX10 | 17728 | -2.808 | 0.0814 | No | ||
| 97 | NDUFS4 | 18111 | -3.233 | 0.0703 | No | ||
| 98 | ATP6V1D | 18360 | -3.491 | 0.0659 | No | ||
| 99 | ATP6V1E1 | 18429 | -3.570 | 0.0698 | No | ||
| 100 | ATP6V0A2 | 19099 | -4.415 | 0.0479 | No | ||
| 101 | ATP6V0C | 19637 | -5.252 | 0.0337 | No | ||
| 102 | NDUFA3 | 19698 | -5.335 | 0.0415 | No | ||
| 103 | ATP6AP1 | 20302 | -6.496 | 0.0268 | No | ||
| 104 | ATP6V1B2 | 20450 | -6.861 | 0.0336 | No | ||
| 105 | ATP6V1H | 20468 | -6.907 | 0.0465 | No | ||
| 106 | ATP6V0D1 | 21421 | -10.719 | 0.0241 | No |