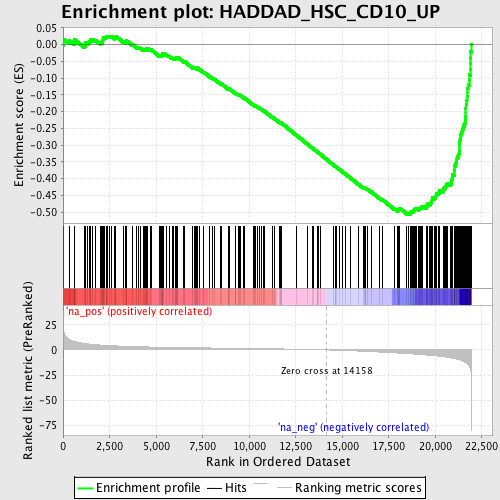
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HADDAD_HSC_CD10_UP |
| Enrichment Score (ES) | -0.5070359 |
| Normalized Enrichment Score (NES) | -2.1665165 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0011715885 |
| FWER p-Value | 0.013 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SOD2 | 24 | 19.385 | 0.0170 | No | ||
| 2 | VDAC1 | 339 | 10.409 | 0.0122 | No | ||
| 3 | SNX2 | 598 | 8.451 | 0.0083 | No | ||
| 4 | GLRX | 613 | 8.366 | 0.0154 | No | ||
| 5 | PSPC1 | 1138 | 6.462 | -0.0027 | No | ||
| 6 | IL7R | 1187 | 6.326 | 0.0010 | No | ||
| 7 | EIF1AY | 1211 | 6.266 | 0.0058 | No | ||
| 8 | SNTB2 | 1303 | 6.030 | 0.0073 | No | ||
| 9 | PCBP2 | 1395 | 5.820 | 0.0085 | No | ||
| 10 | BLK | 1450 | 5.692 | 0.0113 | No | ||
| 11 | UXS1 | 1463 | 5.658 | 0.0161 | No | ||
| 12 | PCDH9 | 1582 | 5.441 | 0.0157 | No | ||
| 13 | CDKN2C | 1747 | 5.171 | 0.0130 | No | ||
| 14 | HIP1R | 2025 | 4.778 | 0.0047 | No | ||
| 15 | TGIF | 2078 | 4.723 | 0.0067 | No | ||
| 16 | HIST1H3D | 2129 | 4.639 | 0.0087 | No | ||
| 17 | HRK | 2132 | 4.634 | 0.0130 | No | ||
| 18 | NELF | 2145 | 4.619 | 0.0167 | No | ||
| 19 | AUTS2 | 2152 | 4.610 | 0.0207 | No | ||
| 20 | PPFIBP1 | 2196 | 4.547 | 0.0230 | No | ||
| 21 | PTPRK | 2304 | 4.414 | 0.0222 | No | ||
| 22 | ZBTB10 | 2332 | 4.379 | 0.0250 | No | ||
| 23 | POU2AF1 | 2407 | 4.304 | 0.0257 | No | ||
| 24 | PCNA | 2502 | 4.211 | 0.0253 | No | ||
| 25 | GNG7 | 2579 | 4.126 | 0.0256 | No | ||
| 26 | P2RX5 | 2771 | 3.970 | 0.0205 | No | ||
| 27 | RHOB | 2798 | 3.952 | 0.0230 | No | ||
| 28 | CTBP2 | 2839 | 3.915 | 0.0248 | No | ||
| 29 | CORO2B | 3244 | 3.621 | 0.0096 | No | ||
| 30 | FMR1 | 3372 | 3.517 | 0.0071 | No | ||
| 31 | CSH2 | 3394 | 3.498 | 0.0094 | No | ||
| 32 | AEBP1 | 3403 | 3.494 | 0.0123 | No | ||
| 33 | HS3ST1 | 3718 | 3.298 | 0.0009 | No | ||
| 34 | TCF4 | 3960 | 3.166 | -0.0073 | No | ||
| 35 | CHD7 | 4026 | 3.129 | -0.0073 | No | ||
| 36 | PHF15 | 4144 | 3.073 | -0.0098 | No | ||
| 37 | IGLL1 | 4296 | 2.996 | -0.0140 | No | ||
| 38 | NPY | 4383 | 2.961 | -0.0152 | No | ||
| 39 | DNASE2B | 4438 | 2.936 | -0.0149 | No | ||
| 40 | MYLK | 4466 | 2.925 | -0.0134 | No | ||
| 41 | GPR125 | 4481 | 2.920 | -0.0114 | No | ||
| 42 | TCF3 | 4510 | 2.909 | -0.0099 | No | ||
| 43 | ATR | 4687 | 2.841 | -0.0154 | No | ||
| 44 | ELL3 | 4741 | 2.817 | -0.0152 | No | ||
| 45 | TOP2A | 5189 | 2.657 | -0.0333 | No | ||
| 46 | ENTPD4 | 5213 | 2.650 | -0.0319 | No | ||
| 47 | HIST1H4H | 5271 | 2.632 | -0.0320 | No | ||
| 48 | ANAPC5 | 5311 | 2.620 | -0.0314 | No | ||
| 49 | HIST1H1E | 5318 | 2.618 | -0.0292 | No | ||
| 50 | EFNB2 | 5332 | 2.614 | -0.0274 | No | ||
| 51 | AKAP12 | 5361 | 2.605 | -0.0262 | No | ||
| 52 | MEF2C | 5401 | 2.593 | -0.0256 | No | ||
| 53 | RCBTB1 | 5556 | 2.541 | -0.0303 | No | ||
| 54 | PKD2 | 5696 | 2.496 | -0.0344 | No | ||
| 55 | PLXNA1 | 5859 | 2.444 | -0.0396 | No | ||
| 56 | CCDC81 | 5930 | 2.419 | -0.0405 | No | ||
| 57 | DDR1 | 6027 | 2.387 | -0.0427 | No | ||
| 58 | SH3TC1 | 6049 | 2.382 | -0.0415 | No | ||
| 59 | CITED2 | 6065 | 2.376 | -0.0399 | No | ||
| 60 | CEP135 | 6071 | 2.374 | -0.0380 | No | ||
| 61 | RRM2 | 6132 | 2.358 | -0.0385 | No | ||
| 62 | SEMA6A | 6142 | 2.353 | -0.0367 | No | ||
| 63 | VPS13A | 6491 | 2.255 | -0.0506 | No | ||
| 64 | COBL | 6542 | 2.241 | -0.0509 | No | ||
| 65 | COL5A1 | 6959 | 2.129 | -0.0680 | No | ||
| 66 | CD72 | 7056 | 2.104 | -0.0705 | No | ||
| 67 | NID2 | 7088 | 2.093 | -0.0699 | No | ||
| 68 | KIF13A | 7148 | 2.079 | -0.0707 | No | ||
| 69 | GPM6B | 7193 | 2.065 | -0.0708 | No | ||
| 70 | REXO2 | 7203 | 2.064 | -0.0693 | No | ||
| 71 | MME | 7343 | 2.025 | -0.0738 | No | ||
| 72 | CAV1 | 7552 | 1.974 | -0.0815 | No | ||
| 73 | P4HA2 | 7889 | 1.885 | -0.0952 | No | ||
| 74 | CD19 | 8029 | 1.852 | -0.0999 | No | ||
| 75 | IRF4 | 8141 | 1.829 | -0.1033 | No | ||
| 76 | SCN3A | 8462 | 1.750 | -0.1164 | No | ||
| 77 | LRMP | 8503 | 1.743 | -0.1166 | No | ||
| 78 | CD79A | 8876 | 1.654 | -0.1322 | No | ||
| 79 | AK1 | 8894 | 1.649 | -0.1314 | No | ||
| 80 | CD24 | 8918 | 1.644 | -0.1309 | No | ||
| 81 | SCHIP1 | 9249 | 1.563 | -0.1447 | No | ||
| 82 | PXDN | 9282 | 1.555 | -0.1447 | No | ||
| 83 | PPP3CC | 9402 | 1.530 | -0.1487 | No | ||
| 84 | PLXNA2 | 9461 | 1.514 | -0.1500 | No | ||
| 85 | MPPED2 | 9533 | 1.496 | -0.1519 | No | ||
| 86 | POU4F1 | 9704 | 1.460 | -0.1583 | No | ||
| 87 | LRP2BP | 9736 | 1.450 | -0.1584 | No | ||
| 88 | RPS11 | 10209 | 1.331 | -0.1789 | No | ||
| 89 | FJX1 | 10307 | 1.310 | -0.1821 | No | ||
| 90 | QRSL1 | 10312 | 1.308 | -0.1811 | No | ||
| 91 | CALM1 | 10454 | 1.273 | -0.1864 | No | ||
| 92 | TMEM118 | 10532 | 1.252 | -0.1888 | No | ||
| 93 | DUSP26 | 10650 | 1.226 | -0.1930 | No | ||
| 94 | MSX1 | 10780 | 1.194 | -0.1978 | No | ||
| 95 | FHIT | 10797 | 1.187 | -0.1974 | No | ||
| 96 | VPREB3 | 11230 | 1.075 | -0.2163 | No | ||
| 97 | CIITA | 11335 | 1.045 | -0.2201 | No | ||
| 98 | BARD1 | 11636 | 0.955 | -0.2330 | No | ||
| 99 | DBN1 | 11665 | 0.946 | -0.2334 | No | ||
| 100 | BLNK | 11697 | 0.938 | -0.2340 | No | ||
| 101 | CD9 | 11727 | 0.926 | -0.2345 | No | ||
| 102 | ALOX5 | 11728 | 0.925 | -0.2336 | No | ||
| 103 | JUN | 12565 | 0.666 | -0.2715 | No | ||
| 104 | MOG | 13135 | 0.463 | -0.2972 | No | ||
| 105 | TAF9B | 13378 | 0.366 | -0.3080 | No | ||
| 106 | FAIM3 | 13443 | 0.340 | -0.3106 | No | ||
| 107 | WFS1 | 13661 | 0.242 | -0.3204 | No | ||
| 108 | IL1A | 13673 | 0.237 | -0.3207 | No | ||
| 109 | GADD45G | 13718 | 0.218 | -0.3225 | No | ||
| 110 | HPS4 | 13816 | 0.163 | -0.3268 | No | ||
| 111 | ALDH5A1 | 14522 | -0.233 | -0.3590 | No | ||
| 112 | MS4A1 | 14610 | -0.274 | -0.3628 | No | ||
| 113 | EFHA1 | 14614 | -0.276 | -0.3626 | No | ||
| 114 | EFHC1 | 14714 | -0.352 | -0.3669 | No | ||
| 115 | MAG | 14840 | -0.439 | -0.3722 | No | ||
| 116 | ZFYVE16 | 15026 | -0.559 | -0.3802 | No | ||
| 117 | TSPAN14 | 15181 | -0.667 | -0.3867 | No | ||
| 118 | STAG3 | 15422 | -0.825 | -0.3969 | No | ||
| 119 | KCNJ2 | 15893 | -1.148 | -0.4175 | No | ||
| 120 | SIAH2 | 16149 | -1.364 | -0.4280 | No | ||
| 121 | P2RY14 | 16194 | -1.404 | -0.4287 | No | ||
| 122 | HIST1H1C | 16221 | -1.424 | -0.4285 | No | ||
| 123 | TCL1A | 16237 | -1.434 | -0.4279 | No | ||
| 124 | BANK1 | 16367 | -1.548 | -0.4324 | No | ||
| 125 | E2F5 | 16561 | -1.721 | -0.4397 | No | ||
| 126 | TLE1 | 17016 | -2.145 | -0.4585 | No | ||
| 127 | PLEKHF2 | 17149 | -2.264 | -0.4625 | No | ||
| 128 | ABCG2 | 17797 | -2.887 | -0.4896 | No | ||
| 129 | AP1B1 | 17979 | -3.080 | -0.4950 | No | ||
| 130 | CRIM1 | 18015 | -3.118 | -0.4937 | No | ||
| 131 | ARID5B | 18018 | -3.120 | -0.4909 | No | ||
| 132 | TIA1 | 18051 | -3.154 | -0.4894 | No | ||
| 133 | SOCS2 | 18085 | -3.196 | -0.4880 | No | ||
| 134 | RIMS3 | 18474 | -3.626 | -0.5025 | No | ||
| 135 | MLXIP | 18567 | -3.720 | -0.5032 | No | ||
| 136 | TOB1 | 18651 | -3.809 | -0.5035 | Yes | ||
| 137 | DNTT | 18678 | -3.852 | -0.5011 | Yes | ||
| 138 | ZHX2 | 18710 | -3.894 | -0.4989 | Yes | ||
| 139 | RB1 | 18763 | -3.958 | -0.4976 | Yes | ||
| 140 | RB1CC1 | 18813 | -4.016 | -0.4961 | Yes | ||
| 141 | ATM | 18851 | -4.077 | -0.4940 | Yes | ||
| 142 | CXCR4 | 18871 | -4.113 | -0.4910 | Yes | ||
| 143 | VPREB1 | 18908 | -4.158 | -0.4888 | Yes | ||
| 144 | EIF2AK3 | 18978 | -4.241 | -0.4880 | Yes | ||
| 145 | SEC22B | 19116 | -4.451 | -0.4902 | Yes | ||
| 146 | WSB2 | 19155 | -4.508 | -0.4877 | Yes | ||
| 147 | SNN | 19215 | -4.604 | -0.4861 | Yes | ||
| 148 | TPD52 | 19277 | -4.695 | -0.4846 | Yes | ||
| 149 | IRS2 | 19314 | -4.756 | -0.4818 | Yes | ||
| 150 | SMAD1 | 19507 | -5.043 | -0.4859 | Yes | ||
| 151 | CACNB3 | 19511 | -5.049 | -0.4813 | Yes | ||
| 152 | DDX3Y | 19556 | -5.134 | -0.4786 | Yes | ||
| 153 | CD79B | 19598 | -5.186 | -0.4756 | Yes | ||
| 154 | MDM1 | 19691 | -5.317 | -0.4749 | Yes | ||
| 155 | ISG20 | 19755 | -5.412 | -0.4727 | Yes | ||
| 156 | TOP2B | 19769 | -5.434 | -0.4683 | Yes | ||
| 157 | ANKRD10 | 19817 | -5.521 | -0.4653 | Yes | ||
| 158 | NGLY1 | 19827 | -5.537 | -0.4605 | Yes | ||
| 159 | JARID1D | 19855 | -5.580 | -0.4566 | Yes | ||
| 160 | CYFIP2 | 19962 | -5.811 | -0.4560 | Yes | ||
| 161 | TNFRSF7 | 20015 | -5.900 | -0.4529 | Yes | ||
| 162 | SLC35D1 | 20037 | -5.953 | -0.4483 | Yes | ||
| 163 | TUBA1 | 20075 | -6.030 | -0.4444 | Yes | ||
| 164 | IRF2 | 20165 | -6.208 | -0.4427 | Yes | ||
| 165 | RAG2 | 20210 | -6.300 | -0.4388 | Yes | ||
| 166 | TRIB2 | 20241 | -6.377 | -0.4343 | Yes | ||
| 167 | ARHGEF7 | 20413 | -6.769 | -0.4358 | Yes | ||
| 168 | ELF2 | 20430 | -6.803 | -0.4302 | Yes | ||
| 169 | SUV420H1 | 20504 | -6.967 | -0.4271 | Yes | ||
| 170 | FOXO1A | 20562 | -7.105 | -0.4231 | Yes | ||
| 171 | TLE4 | 20591 | -7.180 | -0.4177 | Yes | ||
| 172 | USP34 | 20668 | -7.402 | -0.4143 | Yes | ||
| 173 | RBM6 | 20835 | -7.931 | -0.4145 | Yes | ||
| 174 | PRDM2 | 20879 | -8.103 | -0.4089 | Yes | ||
| 175 | PPM1A | 20886 | -8.120 | -0.4016 | Yes | ||
| 176 | PIK3CD | 20906 | -8.191 | -0.3948 | Yes | ||
| 177 | CAPN3 | 20928 | -8.274 | -0.3881 | Yes | ||
| 178 | MAP4K4 | 21016 | -8.644 | -0.3840 | Yes | ||
| 179 | IGF2R | 21017 | -8.646 | -0.3760 | Yes | ||
| 180 | ID3 | 21036 | -8.725 | -0.3686 | Yes | ||
| 181 | NEIL1 | 21037 | -8.727 | -0.3605 | Yes | ||
| 182 | JMJD1C | 21097 | -8.917 | -0.3549 | Yes | ||
| 183 | TTC13 | 21116 | -8.997 | -0.3473 | Yes | ||
| 184 | ITPR1 | 21161 | -9.189 | -0.3408 | Yes | ||
| 185 | RAD52 | 21173 | -9.232 | -0.3327 | Yes | ||
| 186 | DNAJB14 | 21249 | -9.616 | -0.3272 | Yes | ||
| 187 | DACT1 | 21277 | -9.787 | -0.3193 | Yes | ||
| 188 | ZFP36L1 | 21292 | -9.857 | -0.3107 | Yes | ||
| 189 | CDKN1B | 21294 | -9.870 | -0.3016 | Yes | ||
| 190 | EDEM1 | 21303 | -9.918 | -0.2927 | Yes | ||
| 191 | PTPRE | 21324 | -10.022 | -0.2842 | Yes | ||
| 192 | CDC2L5 | 21362 | -10.257 | -0.2764 | Yes | ||
| 193 | TMCC1 | 21366 | -10.274 | -0.2669 | Yes | ||
| 194 | MEF2A | 21429 | -10.782 | -0.2597 | Yes | ||
| 195 | SMARCA4 | 21478 | -11.049 | -0.2516 | Yes | ||
| 196 | ING1 | 21498 | -11.307 | -0.2419 | Yes | ||
| 197 | BTG2 | 21581 | -12.086 | -0.2344 | Yes | ||
| 198 | CD69 | 21606 | -12.413 | -0.2240 | Yes | ||
| 199 | RAG1 | 21632 | -12.680 | -0.2133 | Yes | ||
| 200 | VIL2 | 21638 | -12.815 | -0.2016 | Yes | ||
| 201 | LRIG1 | 21641 | -12.849 | -0.1897 | Yes | ||
| 202 | DYRK2 | 21648 | -12.985 | -0.1778 | Yes | ||
| 203 | PCAF | 21673 | -13.259 | -0.1666 | Yes | ||
| 204 | BTG1 | 21702 | -13.806 | -0.1550 | Yes | ||
| 205 | LAPTM5 | 21712 | -14.002 | -0.1423 | Yes | ||
| 206 | RASGRP1 | 21715 | -14.043 | -0.1293 | Yes | ||
| 207 | STK38 | 21758 | -14.684 | -0.1175 | Yes | ||
| 208 | PPM1B | 21824 | -16.557 | -0.1051 | Yes | ||
| 209 | CENTD1 | 21836 | -16.931 | -0.0898 | Yes | ||
| 210 | LEF1 | 21874 | -18.255 | -0.0745 | Yes | ||
| 211 | BACH2 | 21881 | -18.929 | -0.0571 | Yes | ||
| 212 | VAMP1 | 21887 | -19.080 | -0.0395 | Yes | ||
| 213 | SSBP2 | 21905 | -20.594 | -0.0211 | Yes | ||
| 214 | DCK | 21929 | -24.615 | 0.0008 | Yes |
