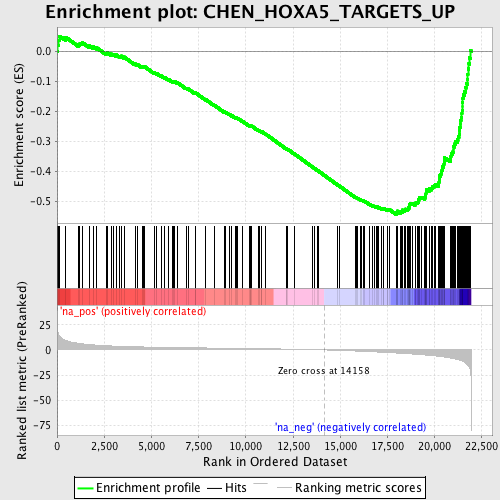
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHEN_HOXA5_TARGETS_UP |
| Enrichment Score (ES) | -0.54430145 |
| Normalized Enrichment Score (NES) | -2.2622147 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.9836604E-4 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTPN2 | 30 | 18.303 | 0.0203 | No | ||
| 2 | CEBPB | 74 | 15.638 | 0.0369 | No | ||
| 3 | SNX5 | 136 | 13.574 | 0.0501 | No | ||
| 4 | SDCCAG1 | 452 | 9.312 | 0.0467 | No | ||
| 5 | PSPC1 | 1138 | 6.462 | 0.0229 | No | ||
| 6 | VEGF | 1209 | 6.268 | 0.0271 | No | ||
| 7 | JOSD3 | 1324 | 5.972 | 0.0290 | No | ||
| 8 | ZRF1 | 1695 | 5.256 | 0.0182 | No | ||
| 9 | CARS | 1906 | 4.931 | 0.0144 | No | ||
| 10 | LIF | 2069 | 4.729 | 0.0126 | No | ||
| 11 | SCYL3 | 2598 | 4.111 | -0.0068 | No | ||
| 12 | NFIL3 | 2646 | 4.055 | -0.0042 | No | ||
| 13 | ASNS | 2861 | 3.899 | -0.0094 | No | ||
| 14 | CREB5 | 2983 | 3.811 | -0.0104 | No | ||
| 15 | RYBP | 3121 | 3.717 | -0.0123 | No | ||
| 16 | RABGGTB | 3303 | 3.577 | -0.0164 | No | ||
| 17 | CASP3 | 3387 | 3.501 | -0.0160 | No | ||
| 18 | ATF3 | 3548 | 3.407 | -0.0193 | No | ||
| 19 | WDR67 | 4133 | 3.080 | -0.0425 | No | ||
| 20 | HLX1 | 4266 | 3.010 | -0.0450 | No | ||
| 21 | CHAC1 | 4501 | 2.913 | -0.0523 | No | ||
| 22 | TNFRSF10B | 4565 | 2.887 | -0.0518 | No | ||
| 23 | PRKRIP1 | 4630 | 2.862 | -0.0513 | No | ||
| 24 | STC2 | 5149 | 2.671 | -0.0719 | No | ||
| 25 | NEK1 | 5268 | 2.633 | -0.0742 | No | ||
| 26 | NUPL1 | 5514 | 2.553 | -0.0824 | No | ||
| 27 | SCG5 | 5693 | 2.497 | -0.0877 | No | ||
| 28 | ARIH1 | 5888 | 2.435 | -0.0937 | No | ||
| 29 | ZCCHC10 | 6135 | 2.356 | -0.1022 | No | ||
| 30 | DPP8 | 6143 | 2.353 | -0.0997 | No | ||
| 31 | CARF | 6215 | 2.331 | -0.1002 | No | ||
| 32 | TRPC1 | 6358 | 2.289 | -0.1040 | No | ||
| 33 | TMEFF1 | 6830 | 2.161 | -0.1231 | No | ||
| 34 | FOSB | 6942 | 2.132 | -0.1257 | No | ||
| 35 | TBX3 | 7307 | 2.036 | -0.1400 | No | ||
| 36 | ING3 | 7332 | 2.028 | -0.1387 | No | ||
| 37 | NOC3L | 7866 | 1.890 | -0.1609 | No | ||
| 38 | DSCR1 | 8326 | 1.782 | -0.1798 | No | ||
| 39 | DUSP8 | 8849 | 1.661 | -0.2018 | No | ||
| 40 | GAD1 | 8925 | 1.642 | -0.2033 | No | ||
| 41 | SOX30 | 9126 | 1.594 | -0.2106 | No | ||
| 42 | CEP76 | 9262 | 1.561 | -0.2150 | No | ||
| 43 | BDKRB1 | 9458 | 1.515 | -0.2221 | No | ||
| 44 | CTH | 9494 | 1.507 | -0.2220 | No | ||
| 45 | DCLRE1C | 9559 | 1.491 | -0.2231 | No | ||
| 46 | SNX16 | 9839 | 1.426 | -0.2343 | No | ||
| 47 | SNAI2 | 10175 | 1.340 | -0.2480 | No | ||
| 48 | NFKBIE | 10229 | 1.326 | -0.2489 | No | ||
| 49 | PPIG | 10237 | 1.325 | -0.2477 | No | ||
| 50 | CENPC1 | 10306 | 1.310 | -0.2492 | No | ||
| 51 | HRH1 | 10660 | 1.224 | -0.2640 | No | ||
| 52 | PTN | 10739 | 1.206 | -0.2661 | No | ||
| 53 | SPATA5L1 | 10822 | 1.182 | -0.2685 | No | ||
| 54 | ZZZ3 | 10839 | 1.178 | -0.2678 | No | ||
| 55 | TPTE | 11062 | 1.118 | -0.2767 | No | ||
| 56 | DLX2 | 12161 | 0.794 | -0.3262 | No | ||
| 57 | PPWD1 | 12173 | 0.791 | -0.3258 | No | ||
| 58 | FOSL2 | 12202 | 0.782 | -0.3261 | No | ||
| 59 | JUN | 12565 | 0.666 | -0.3419 | No | ||
| 60 | KIN | 13517 | 0.309 | -0.3852 | No | ||
| 61 | ZNF35 | 13655 | 0.243 | -0.3912 | No | ||
| 62 | PMAIP1 | 13796 | 0.171 | -0.3975 | No | ||
| 63 | NSUN6 | 13842 | 0.152 | -0.3993 | No | ||
| 64 | F3 | 14869 | -0.457 | -0.4459 | No | ||
| 65 | PTGS2 | 14970 | -0.509 | -0.4499 | No | ||
| 66 | KLF5 | 15788 | -1.070 | -0.4861 | No | ||
| 67 | TAF1A | 15839 | -1.115 | -0.4871 | No | ||
| 68 | SEC31A | 15943 | -1.194 | -0.4904 | No | ||
| 69 | KLF10 | 16085 | -1.313 | -0.4954 | No | ||
| 70 | RIPK2 | 16143 | -1.360 | -0.4964 | No | ||
| 71 | AGTPBP1 | 16216 | -1.419 | -0.4980 | No | ||
| 72 | SEC24A | 16309 | -1.496 | -0.5004 | No | ||
| 73 | GEM | 16556 | -1.714 | -0.5097 | No | ||
| 74 | PTBP2 | 16729 | -1.863 | -0.5154 | No | ||
| 75 | FBXO38 | 16800 | -1.929 | -0.5163 | No | ||
| 76 | RBM15 | 16907 | -2.037 | -0.5188 | No | ||
| 77 | RIC8B | 16953 | -2.076 | -0.5184 | No | ||
| 78 | THUMPD2 | 17055 | -2.183 | -0.5204 | No | ||
| 79 | ZNF3 | 17196 | -2.298 | -0.5241 | No | ||
| 80 | KIF18A | 17283 | -2.370 | -0.5253 | No | ||
| 81 | SFRS12 | 17306 | -2.399 | -0.5234 | No | ||
| 82 | SMCHD1 | 17489 | -2.593 | -0.5287 | No | ||
| 83 | CAB39 | 17508 | -2.607 | -0.5265 | No | ||
| 84 | TANK | 17608 | -2.694 | -0.5278 | No | ||
| 85 | FEM1C | 17968 | -3.072 | -0.5407 | Yes | ||
| 86 | CDK7 | 18002 | -3.105 | -0.5385 | Yes | ||
| 87 | DAAM1 | 18036 | -3.135 | -0.5363 | Yes | ||
| 88 | TUFT1 | 18038 | -3.136 | -0.5326 | Yes | ||
| 89 | RBBP6 | 18221 | -3.350 | -0.5370 | Yes | ||
| 90 | GPATC2 | 18254 | -3.387 | -0.5345 | Yes | ||
| 91 | ADNP | 18312 | -3.434 | -0.5330 | Yes | ||
| 92 | EIF5 | 18319 | -3.440 | -0.5292 | Yes | ||
| 93 | DUSP5 | 18425 | -3.567 | -0.5298 | Yes | ||
| 94 | SETDB1 | 18435 | -3.575 | -0.5260 | Yes | ||
| 95 | CLK1 | 18579 | -3.733 | -0.5281 | Yes | ||
| 96 | BACH1 | 18602 | -3.759 | -0.5247 | Yes | ||
| 97 | TSC22D1 | 18629 | -3.788 | -0.5214 | Yes | ||
| 98 | RCHY1 | 18661 | -3.823 | -0.5183 | Yes | ||
| 99 | TAF2 | 18679 | -3.852 | -0.5145 | Yes | ||
| 100 | DUSP6 | 18680 | -3.854 | -0.5099 | Yes | ||
| 101 | TBK1 | 18737 | -3.926 | -0.5079 | Yes | ||
| 102 | CCNL1 | 18828 | -4.039 | -0.5072 | Yes | ||
| 103 | SAT1 | 18967 | -4.230 | -0.5085 | Yes | ||
| 104 | CNOT2 | 19015 | -4.303 | -0.5056 | Yes | ||
| 105 | KLF6 | 19095 | -4.409 | -0.5040 | Yes | ||
| 106 | IFRD1 | 19138 | -4.480 | -0.5006 | Yes | ||
| 107 | MARK3 | 19140 | -4.484 | -0.4954 | Yes | ||
| 108 | FOS | 19178 | -4.546 | -0.4917 | Yes | ||
| 109 | JMJD1A | 19185 | -4.554 | -0.4866 | Yes | ||
| 110 | FNBP4 | 19304 | -4.732 | -0.4864 | Yes | ||
| 111 | FOXO3A | 19458 | -4.980 | -0.4875 | Yes | ||
| 112 | MAFF | 19502 | -5.036 | -0.4835 | Yes | ||
| 113 | IRF7 | 19524 | -5.076 | -0.4784 | Yes | ||
| 114 | SLTM | 19546 | -5.114 | -0.4734 | Yes | ||
| 115 | RLF | 19551 | -5.128 | -0.4675 | Yes | ||
| 116 | BIRC2 | 19566 | -5.145 | -0.4620 | Yes | ||
| 117 | LIG4 | 19712 | -5.351 | -0.4623 | Yes | ||
| 118 | CLK4 | 19723 | -5.369 | -0.4564 | Yes | ||
| 119 | IER2 | 19863 | -5.603 | -0.4562 | Yes | ||
| 120 | ZCCHC8 | 19907 | -5.676 | -0.4514 | Yes | ||
| 121 | PPP1R12A | 19974 | -5.824 | -0.4476 | Yes | ||
| 122 | CREM | 20025 | -5.922 | -0.4428 | Yes | ||
| 123 | ARID4B | 20206 | -6.287 | -0.4436 | Yes | ||
| 124 | BCL10 | 20231 | -6.352 | -0.4372 | Yes | ||
| 125 | CEBPG | 20240 | -6.372 | -0.4300 | Yes | ||
| 126 | ARNTL | 20247 | -6.382 | -0.4228 | Yes | ||
| 127 | MORC3 | 20258 | -6.408 | -0.4156 | Yes | ||
| 128 | INTS6 | 20327 | -6.551 | -0.4110 | Yes | ||
| 129 | CNOT4 | 20347 | -6.588 | -0.4041 | Yes | ||
| 130 | STRN3 | 20390 | -6.698 | -0.3980 | Yes | ||
| 131 | RELB | 20410 | -6.758 | -0.3909 | Yes | ||
| 132 | TNFRSF9 | 20436 | -6.820 | -0.3840 | Yes | ||
| 133 | TIPARP | 20496 | -6.955 | -0.3784 | Yes | ||
| 134 | SUV420H1 | 20504 | -6.967 | -0.3705 | Yes | ||
| 135 | CCNT2 | 20516 | -6.990 | -0.3627 | Yes | ||
| 136 | CHD9 | 20525 | -7.011 | -0.3548 | Yes | ||
| 137 | WAC | 20840 | -7.951 | -0.3598 | Yes | ||
| 138 | ZCCHC6 | 20866 | -8.034 | -0.3514 | Yes | ||
| 139 | POLR3F | 20876 | -8.098 | -0.3422 | Yes | ||
| 140 | AKAP10 | 20957 | -8.399 | -0.3360 | Yes | ||
| 141 | PRPF38B | 20988 | -8.539 | -0.3272 | Yes | ||
| 142 | PRPF3 | 20990 | -8.545 | -0.3172 | Yes | ||
| 143 | RBM39 | 21045 | -8.749 | -0.3093 | Yes | ||
| 144 | JMJD1C | 21097 | -8.917 | -0.3010 | Yes | ||
| 145 | TSC22D2 | 21207 | -9.406 | -0.2949 | Yes | ||
| 146 | DMTF1 | 21245 | -9.609 | -0.2852 | Yes | ||
| 147 | JUNB | 21298 | -9.897 | -0.2759 | Yes | ||
| 148 | MCL1 | 21326 | -10.029 | -0.2652 | Yes | ||
| 149 | PGS1 | 21339 | -10.118 | -0.2538 | Yes | ||
| 150 | EGR1 | 21359 | -10.247 | -0.2425 | Yes | ||
| 151 | RBM5 | 21367 | -10.275 | -0.2307 | Yes | ||
| 152 | GADD45B | 21406 | -10.572 | -0.2199 | Yes | ||
| 153 | SFPQ | 21450 | -10.874 | -0.2090 | Yes | ||
| 154 | SUHW4 | 21457 | -10.946 | -0.1963 | Yes | ||
| 155 | RNF111 | 21480 | -11.117 | -0.1842 | Yes | ||
| 156 | TNFAIP3 | 21482 | -11.148 | -0.1710 | Yes | ||
| 157 | NCOA3 | 21501 | -11.314 | -0.1584 | Yes | ||
| 158 | BIRC3 | 21517 | -11.412 | -0.1456 | Yes | ||
| 159 | MARCH7 | 21597 | -12.301 | -0.1346 | Yes | ||
| 160 | TLK2 | 21642 | -12.867 | -0.1214 | Yes | ||
| 161 | PELI1 | 21674 | -13.286 | -0.1071 | Yes | ||
| 162 | IHPK2 | 21750 | -14.578 | -0.0933 | Yes | ||
| 163 | ETS2 | 21763 | -14.814 | -0.0763 | Yes | ||
| 164 | NFKBIA | 21775 | -15.114 | -0.0589 | Yes | ||
| 165 | NEDD9 | 21820 | -16.353 | -0.0415 | Yes | ||
| 166 | PCTK2 | 21857 | -17.748 | -0.0221 | Yes | ||
| 167 | CRY1 | 21920 | -22.141 | 0.0012 | Yes |
