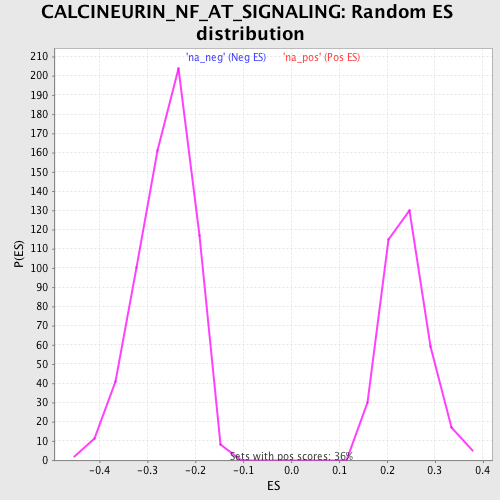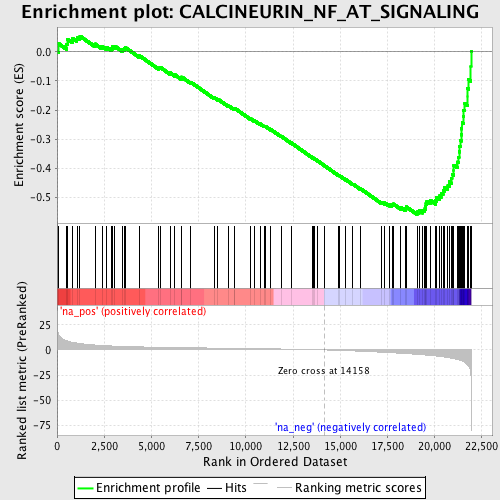
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CALCINEURIN_NF_AT_SIGNALING |
| Enrichment Score (ES) | -0.55903035 |
| Normalized Enrichment Score (NES) | -2.1362984 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0012734932 |
| FWER p-Value | 0.018 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 74 | 15.638 | 0.0282 | No | ||
| 2 | HRAS | 510 | 8.966 | 0.0265 | No | ||
| 3 | NCK2 | 543 | 8.728 | 0.0427 | No | ||
| 4 | CSNK2B | 803 | 7.574 | 0.0461 | No | ||
| 5 | FCGR3A | 1053 | 6.726 | 0.0483 | No | ||
| 6 | VEGF | 1209 | 6.268 | 0.0539 | No | ||
| 7 | BAD | 2011 | 4.790 | 0.0270 | No | ||
| 8 | NFATC2 | 2387 | 4.322 | 0.0185 | No | ||
| 9 | P2RX7 | 2631 | 4.076 | 0.0157 | No | ||
| 10 | XPO5 | 2886 | 3.880 | 0.0119 | No | ||
| 11 | CDKN1A | 2944 | 3.835 | 0.0171 | No | ||
| 12 | IL3 | 3065 | 3.756 | 0.0192 | No | ||
| 13 | FOSL1 | 3479 | 3.447 | 0.0072 | No | ||
| 14 | IL2RA | 3564 | 3.387 | 0.0102 | No | ||
| 15 | PIN1 | 3616 | 3.352 | 0.0147 | No | ||
| 16 | FKBP1B | 4372 | 2.965 | -0.0139 | No | ||
| 17 | NFATC4 | 5397 | 2.593 | -0.0555 | No | ||
| 18 | CAMK2B | 5454 | 2.573 | -0.0528 | No | ||
| 19 | OPRD1 | 5988 | 2.400 | -0.0724 | No | ||
| 20 | IFNG | 6243 | 2.320 | -0.0793 | No | ||
| 21 | CNR1 | 6598 | 2.225 | -0.0910 | No | ||
| 22 | GATA4 | 6600 | 2.225 | -0.0865 | No | ||
| 23 | IL10 | 7078 | 2.096 | -0.1041 | No | ||
| 24 | VAV3 | 8314 | 1.784 | -0.1570 | No | ||
| 25 | TRPV6 | 8515 | 1.739 | -0.1626 | No | ||
| 26 | IFNA1 | 9097 | 1.601 | -0.1860 | No | ||
| 27 | PPP3CC | 9402 | 1.530 | -0.1968 | No | ||
| 28 | NFKBIB | 9422 | 1.525 | -0.1946 | No | ||
| 29 | NFKBIE | 10229 | 1.326 | -0.2288 | No | ||
| 30 | CALM1 | 10454 | 1.273 | -0.2364 | No | ||
| 31 | RPL13A | 10754 | 1.199 | -0.2477 | No | ||
| 32 | FCER1A | 10985 | 1.140 | -0.2559 | No | ||
| 33 | MEF2B | 11030 | 1.125 | -0.2556 | No | ||
| 34 | NPPB | 11325 | 1.048 | -0.2670 | No | ||
| 35 | CALM2 | 11871 | 0.882 | -0.2901 | No | ||
| 36 | IL4 | 12402 | 0.724 | -0.3129 | No | ||
| 37 | IL13 | 13554 | 0.294 | -0.3650 | No | ||
| 38 | CSF2 | 13564 | 0.288 | -0.3648 | No | ||
| 39 | IL6 | 13644 | 0.251 | -0.3679 | No | ||
| 40 | MYF5 | 13806 | 0.168 | -0.3749 | No | ||
| 41 | SFN | 14174 | -0.015 | -0.3917 | No | ||
| 42 | IFNB1 | 14917 | -0.487 | -0.4247 | No | ||
| 43 | GSK3A | 14949 | -0.501 | -0.4251 | No | ||
| 44 | PPIA | 15294 | -0.732 | -0.4393 | No | ||
| 45 | IL1B | 15675 | -1.000 | -0.4547 | No | ||
| 46 | TNF | 16079 | -1.307 | -0.4705 | No | ||
| 47 | MAPK9 | 17214 | -2.310 | -0.5177 | No | ||
| 48 | CALM3 | 17354 | -2.438 | -0.5191 | No | ||
| 49 | NUP214 | 17596 | -2.687 | -0.5247 | No | ||
| 50 | PAK1 | 17746 | -2.826 | -0.5258 | No | ||
| 51 | MAPK14 | 17811 | -2.903 | -0.5229 | No | ||
| 52 | PPP3R1 | 18216 | -3.345 | -0.5346 | No | ||
| 53 | CABIN1 | 18461 | -3.612 | -0.5384 | No | ||
| 54 | GRLF1 | 18502 | -3.660 | -0.5329 | No | ||
| 55 | CTLA4 | 19075 | -4.385 | -0.5502 | Yes | ||
| 56 | FOS | 19178 | -4.546 | -0.5456 | Yes | ||
| 57 | CD3G | 19364 | -4.837 | -0.5443 | Yes | ||
| 58 | NFKB2 | 19492 | -5.020 | -0.5400 | Yes | ||
| 59 | PTPRC | 19517 | -5.062 | -0.5308 | Yes | ||
| 60 | NFAT5 | 19545 | -5.113 | -0.5217 | Yes | ||
| 61 | ACTB | 19594 | -5.181 | -0.5134 | Yes | ||
| 62 | CAMK4 | 19772 | -5.438 | -0.5105 | Yes | ||
| 63 | VAV1 | 20066 | -6.018 | -0.5118 | Yes | ||
| 64 | GATA3 | 20113 | -6.093 | -0.5016 | Yes | ||
| 65 | MAP2K7 | 20261 | -6.417 | -0.4953 | Yes | ||
| 66 | PPP3CB | 20348 | -6.597 | -0.4859 | Yes | ||
| 67 | VAV2 | 20452 | -6.862 | -0.4767 | Yes | ||
| 68 | SP3 | 20510 | -6.981 | -0.4652 | Yes | ||
| 69 | ICOS | 20711 | -7.531 | -0.4591 | Yes | ||
| 70 | EP300 | 20781 | -7.751 | -0.4466 | Yes | ||
| 71 | TGFB1 | 20897 | -8.163 | -0.4354 | Yes | ||
| 72 | CSNK2A1 | 20926 | -8.269 | -0.4199 | Yes | ||
| 73 | TRAF2 | 21018 | -8.648 | -0.4066 | Yes | ||
| 74 | RELA | 21022 | -8.671 | -0.3892 | Yes | ||
| 75 | MAPK8 | 21208 | -9.408 | -0.3786 | Yes | ||
| 76 | SLA | 21278 | -9.796 | -0.3620 | Yes | ||
| 77 | JUNB | 21298 | -9.897 | -0.3428 | Yes | ||
| 78 | ITK | 21334 | -10.083 | -0.3241 | Yes | ||
| 79 | BCL2 | 21371 | -10.300 | -0.3049 | Yes | ||
| 80 | CREBBP | 21411 | -10.605 | -0.2852 | Yes | ||
| 81 | MEF2A | 21429 | -10.782 | -0.2642 | Yes | ||
| 82 | GSK3B | 21455 | -10.935 | -0.2432 | Yes | ||
| 83 | NFATC1 | 21534 | -11.557 | -0.2234 | Yes | ||
| 84 | CD3E | 21545 | -11.647 | -0.2003 | Yes | ||
| 85 | CD69 | 21606 | -12.413 | -0.1779 | Yes | ||
| 86 | MEF2D | 21742 | -14.522 | -0.1548 | Yes | ||
| 87 | NFATC3 | 21743 | -14.538 | -0.1254 | Yes | ||
| 88 | SP1 | 21811 | -15.984 | -0.0961 | Yes | ||
| 89 | EGR3 | 21930 | -24.862 | -0.0512 | Yes | ||
| 90 | EGR2 | 21933 | -25.685 | 0.0006 | Yes |