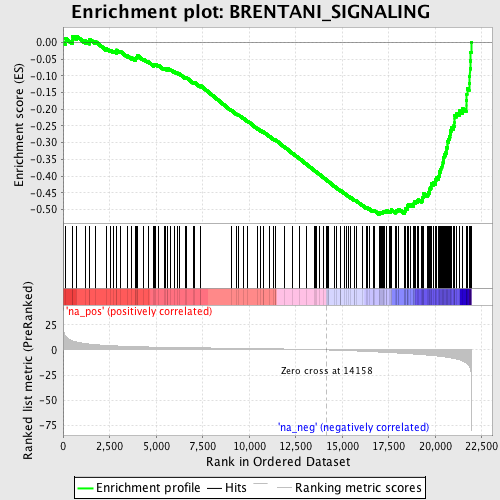
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BRENTANI_SIGNALING |
| Enrichment Score (ES) | -0.51495886 |
| Normalized Enrichment Score (NES) | -2.1389685 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.001485742 |
| FWER p-Value | 0.018 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAN | 122 | 13.946 | 0.0127 | No | ||
| 2 | SMO | 494 | 9.076 | 0.0075 | No | ||
| 3 | HRAS | 510 | 8.966 | 0.0186 | No | ||
| 4 | CRK | 726 | 7.882 | 0.0191 | No | ||
| 5 | ITGA9 | 1216 | 6.259 | 0.0048 | No | ||
| 6 | MAPK1 | 1423 | 5.768 | 0.0029 | No | ||
| 7 | IL2RB | 1434 | 5.742 | 0.0100 | No | ||
| 8 | WNT4 | 1741 | 5.178 | 0.0027 | No | ||
| 9 | PTPRK | 2304 | 4.414 | -0.0173 | No | ||
| 10 | PTCH2 | 2547 | 4.156 | -0.0230 | No | ||
| 11 | MAPKAPK2 | 2720 | 3.998 | -0.0256 | No | ||
| 12 | MAPK12 | 2869 | 3.894 | -0.0273 | No | ||
| 13 | NOTCH4 | 2875 | 3.889 | -0.0225 | No | ||
| 14 | JAG2 | 3061 | 3.758 | -0.0260 | No | ||
| 15 | TTK | 3448 | 3.461 | -0.0392 | No | ||
| 16 | ITGA7 | 3679 | 3.319 | -0.0454 | No | ||
| 17 | TYRO3 | 3877 | 3.216 | -0.0503 | No | ||
| 18 | ITGB1 | 3903 | 3.201 | -0.0472 | No | ||
| 19 | NOTCH3 | 3928 | 3.185 | -0.0441 | No | ||
| 20 | CXCL12 | 3981 | 3.157 | -0.0424 | No | ||
| 21 | BTK | 3984 | 3.157 | -0.0383 | No | ||
| 22 | MAP2K3 | 4337 | 2.980 | -0.0506 | No | ||
| 23 | TNFRSF10B | 4565 | 2.887 | -0.0572 | No | ||
| 24 | MCC | 4876 | 2.764 | -0.0678 | No | ||
| 25 | CXCL1 | 4897 | 2.757 | -0.0651 | No | ||
| 26 | ITGA3 | 4977 | 2.731 | -0.0652 | No | ||
| 27 | GRB14 | 5140 | 2.676 | -0.0691 | No | ||
| 28 | MAPK10 | 5422 | 2.586 | -0.0786 | No | ||
| 29 | ARHGEF12 | 5484 | 2.562 | -0.0781 | No | ||
| 30 | MAP2K5 | 5603 | 2.524 | -0.0802 | No | ||
| 31 | ITGB5 | 5632 | 2.516 | -0.0782 | No | ||
| 32 | SOS1 | 5789 | 2.466 | -0.0821 | No | ||
| 33 | SUFU | 5983 | 2.403 | -0.0878 | No | ||
| 34 | JAG1 | 6125 | 2.359 | -0.0912 | No | ||
| 35 | IFNG | 6243 | 2.320 | -0.0935 | No | ||
| 36 | MAPK4 | 6559 | 2.234 | -0.1051 | No | ||
| 37 | WNT5A | 6638 | 2.213 | -0.1058 | No | ||
| 38 | ERG | 7011 | 2.116 | -0.1201 | No | ||
| 39 | IL10 | 7078 | 2.096 | -0.1204 | No | ||
| 40 | ITGAX | 7364 | 2.021 | -0.1308 | No | ||
| 41 | ARHGAP6 | 7368 | 2.020 | -0.1283 | No | ||
| 42 | CXCL2 | 9039 | 1.616 | -0.2029 | No | ||
| 43 | ITGA2 | 9328 | 1.545 | -0.2141 | No | ||
| 44 | MAP3K4 | 9437 | 1.520 | -0.2170 | No | ||
| 45 | IL15 | 9679 | 1.466 | -0.2262 | No | ||
| 46 | BCR | 9930 | 1.403 | -0.2358 | No | ||
| 47 | CALM1 | 10454 | 1.273 | -0.2582 | No | ||
| 48 | ITGA1 | 10623 | 1.232 | -0.2643 | No | ||
| 49 | ABL2 | 10761 | 1.198 | -0.2690 | No | ||
| 50 | MAPK13 | 10765 | 1.197 | -0.2676 | No | ||
| 51 | GRB7 | 11069 | 1.117 | -0.2800 | No | ||
| 52 | ITGAM | 11326 | 1.048 | -0.2904 | No | ||
| 53 | IQGAP2 | 11409 | 1.021 | -0.2928 | No | ||
| 54 | IGFBP3 | 11414 | 1.019 | -0.2917 | No | ||
| 55 | VIPR1 | 11920 | 0.864 | -0.3137 | No | ||
| 56 | TNFSF4 | 12334 | 0.745 | -0.3317 | No | ||
| 57 | NF1 | 12700 | 0.627 | -0.3477 | No | ||
| 58 | WNT1 | 13064 | 0.489 | -0.3637 | No | ||
| 59 | WNT2 | 13510 | 0.312 | -0.3837 | No | ||
| 60 | IL13 | 13554 | 0.294 | -0.3853 | No | ||
| 61 | RAP1A | 13631 | 0.258 | -0.3885 | No | ||
| 62 | ROS1 | 13801 | 0.169 | -0.3960 | No | ||
| 63 | IGFBP1 | 13986 | 0.088 | -0.4043 | No | ||
| 64 | SFN | 14174 | -0.015 | -0.4129 | No | ||
| 65 | ARHGEF16 | 14196 | -0.029 | -0.4138 | No | ||
| 66 | INHA | 14268 | -0.072 | -0.4170 | No | ||
| 67 | MYD88 | 14281 | -0.077 | -0.4174 | No | ||
| 68 | ITGB4 | 14582 | -0.265 | -0.4309 | No | ||
| 69 | IL18R1 | 14703 | -0.349 | -0.4359 | No | ||
| 70 | MPL | 14900 | -0.475 | -0.4443 | No | ||
| 71 | REL | 14913 | -0.486 | -0.4442 | No | ||
| 72 | IFNB1 | 14917 | -0.487 | -0.4437 | No | ||
| 73 | ITGB8 | 15102 | -0.614 | -0.4514 | No | ||
| 74 | SRC | 15241 | -0.705 | -0.4568 | No | ||
| 75 | IL12RB1 | 15346 | -0.770 | -0.4605 | No | ||
| 76 | AXL | 15466 | -0.856 | -0.4649 | No | ||
| 77 | IRS1 | 15669 | -0.991 | -0.4729 | No | ||
| 78 | IL1B | 15675 | -1.000 | -0.4718 | No | ||
| 79 | MAP2K1 | 15741 | -1.040 | -0.4734 | No | ||
| 80 | TNF | 16079 | -1.307 | -0.4872 | No | ||
| 81 | MUSK | 16304 | -1.493 | -0.4955 | No | ||
| 82 | ITGA4 | 16331 | -1.513 | -0.4947 | No | ||
| 83 | ZAP70 | 16444 | -1.608 | -0.4977 | No | ||
| 84 | TNFSF13 | 16654 | -1.800 | -0.5050 | No | ||
| 85 | CTNNB1 | 16702 | -1.841 | -0.5047 | No | ||
| 86 | SHC1 | 16723 | -1.857 | -0.5032 | No | ||
| 87 | TRAF6 | 16980 | -2.105 | -0.5122 | Yes | ||
| 88 | MAPK7 | 16999 | -2.122 | -0.5102 | Yes | ||
| 89 | AKT1 | 17066 | -2.193 | -0.5104 | Yes | ||
| 90 | TYK2 | 17095 | -2.213 | -0.5088 | Yes | ||
| 91 | NCK1 | 17148 | -2.262 | -0.5082 | Yes | ||
| 92 | MAP3K7IP1 | 17192 | -2.295 | -0.5072 | Yes | ||
| 93 | MAPK9 | 17214 | -2.310 | -0.5051 | Yes | ||
| 94 | TIAM1 | 17271 | -2.359 | -0.5046 | Yes | ||
| 95 | MAP3K14 | 17377 | -2.462 | -0.5062 | Yes | ||
| 96 | ITGAV | 17397 | -2.484 | -0.5038 | Yes | ||
| 97 | RGS16 | 17520 | -2.621 | -0.5060 | Yes | ||
| 98 | TANK | 17608 | -2.694 | -0.5064 | Yes | ||
| 99 | MAP2K6 | 17632 | -2.723 | -0.5039 | Yes | ||
| 100 | ITGB7 | 17644 | -2.731 | -0.5008 | Yes | ||
| 101 | MAP3K8 | 17860 | -2.954 | -0.5068 | Yes | ||
| 102 | ITGA5 | 17889 | -2.987 | -0.5042 | Yes | ||
| 103 | JAK3 | 17929 | -3.030 | -0.5020 | Yes | ||
| 104 | DGKQ | 17999 | -3.103 | -0.5011 | Yes | ||
| 105 | MAPK11 | 18040 | -3.143 | -0.4989 | Yes | ||
| 106 | BRAF | 18334 | -3.456 | -0.5078 | Yes | ||
| 107 | MKNK1 | 18365 | -3.498 | -0.5046 | Yes | ||
| 108 | TNFRSF1B | 18378 | -3.515 | -0.5005 | Yes | ||
| 109 | IKBKG | 18397 | -3.530 | -0.4967 | Yes | ||
| 110 | ARHGAP1 | 18490 | -3.648 | -0.4962 | Yes | ||
| 111 | SYK | 18493 | -3.653 | -0.4915 | Yes | ||
| 112 | ILK | 18512 | -3.670 | -0.4875 | Yes | ||
| 113 | AXIN2 | 18561 | -3.716 | -0.4848 | Yes | ||
| 114 | RALBP1 | 18685 | -3.861 | -0.4854 | Yes | ||
| 115 | TNFRSF14 | 18820 | -4.030 | -0.4863 | Yes | ||
| 116 | ATM | 18851 | -4.077 | -0.4823 | Yes | ||
| 117 | PIK3C3 | 18859 | -4.098 | -0.4772 | Yes | ||
| 118 | IFNAR2 | 18945 | -4.206 | -0.4756 | Yes | ||
| 119 | ITGB3 | 19031 | -4.324 | -0.4739 | Yes | ||
| 120 | RAF1 | 19074 | -4.383 | -0.4701 | Yes | ||
| 121 | PTEN | 19245 | -4.642 | -0.4718 | Yes | ||
| 122 | IRS2 | 19314 | -4.756 | -0.4687 | Yes | ||
| 123 | MAP3K12 | 19326 | -4.778 | -0.4629 | Yes | ||
| 124 | GNB1 | 19355 | -4.818 | -0.4579 | Yes | ||
| 125 | STAT6 | 19356 | -4.818 | -0.4516 | Yes | ||
| 126 | BIRC2 | 19566 | -5.145 | -0.4544 | Yes | ||
| 127 | AXIN1 | 19618 | -5.227 | -0.4499 | Yes | ||
| 128 | STAT5A | 19672 | -5.285 | -0.4454 | Yes | ||
| 129 | JAK1 | 19675 | -5.293 | -0.4386 | Yes | ||
| 130 | ABL1 | 19742 | -5.391 | -0.4345 | Yes | ||
| 131 | CAMK4 | 19772 | -5.438 | -0.4287 | Yes | ||
| 132 | APC | 19779 | -5.445 | -0.4219 | Yes | ||
| 133 | SH3BP2 | 19894 | -5.655 | -0.4197 | Yes | ||
| 134 | ROCK1 | 20004 | -5.882 | -0.4170 | Yes | ||
| 135 | MAS1 | 20024 | -5.920 | -0.4101 | Yes | ||
| 136 | VAV1 | 20066 | -6.018 | -0.4041 | Yes | ||
| 137 | DGKG | 20188 | -6.246 | -0.4015 | Yes | ||
| 138 | AKT2 | 20209 | -6.297 | -0.3941 | Yes | ||
| 139 | BCL10 | 20231 | -6.352 | -0.3868 | Yes | ||
| 140 | MAP2K7 | 20261 | -6.417 | -0.3797 | Yes | ||
| 141 | ITGB2 | 20306 | -6.502 | -0.3732 | Yes | ||
| 142 | MAP2K2 | 20359 | -6.618 | -0.3669 | Yes | ||
| 143 | RELB | 20410 | -6.758 | -0.3603 | Yes | ||
| 144 | MAP3K3 | 20422 | -6.791 | -0.3519 | Yes | ||
| 145 | SKIL | 20440 | -6.832 | -0.3438 | Yes | ||
| 146 | STAT2 | 20472 | -6.911 | -0.3361 | Yes | ||
| 147 | CRKL | 20555 | -7.087 | -0.3306 | Yes | ||
| 148 | MAPKAPK5 | 20596 | -7.204 | -0.3230 | Yes | ||
| 149 | ITGA6 | 20601 | -7.218 | -0.3137 | Yes | ||
| 150 | AKAP13 | 20654 | -7.366 | -0.3064 | Yes | ||
| 151 | ITGAE | 20655 | -7.369 | -0.2968 | Yes | ||
| 152 | DLG1 | 20702 | -7.506 | -0.2890 | Yes | ||
| 153 | IFNGR1 | 20745 | -7.609 | -0.2810 | Yes | ||
| 154 | STAT3 | 20789 | -7.787 | -0.2728 | Yes | ||
| 155 | RASA1 | 20833 | -7.927 | -0.2644 | Yes | ||
| 156 | TRAF3 | 20854 | -7.990 | -0.2548 | Yes | ||
| 157 | TRAF1 | 20992 | -8.556 | -0.2499 | Yes | ||
| 158 | MAP4K4 | 21016 | -8.644 | -0.2396 | Yes | ||
| 159 | RELA | 21022 | -8.671 | -0.2285 | Yes | ||
| 160 | PIK3CG | 21024 | -8.675 | -0.2171 | Yes | ||
| 161 | NRAS | 21157 | -9.167 | -0.2112 | Yes | ||
| 162 | FYN | 21301 | -9.911 | -0.2047 | Yes | ||
| 163 | MAP2K4 | 21466 | -10.991 | -0.1979 | Yes | ||
| 164 | STAT5B | 21655 | -13.076 | -0.1894 | Yes | ||
| 165 | STAT1 | 21656 | -13.091 | -0.1722 | Yes | ||
| 166 | PTK2B | 21693 | -13.678 | -0.1559 | Yes | ||
| 167 | TNFRSF1A | 21703 | -13.816 | -0.1382 | Yes | ||
| 168 | YWHAE | 21814 | -16.134 | -0.1221 | Yes | ||
| 169 | IFNAR1 | 21831 | -16.830 | -0.1008 | Yes | ||
| 170 | TNFRSF4 | 21863 | -17.875 | -0.0788 | Yes | ||
| 171 | PDPK1 | 21879 | -18.696 | -0.0550 | Yes | ||
| 172 | IL6ST | 21897 | -19.754 | -0.0299 | Yes | ||
| 173 | DGKA | 21928 | -24.490 | 0.0009 | Yes |
