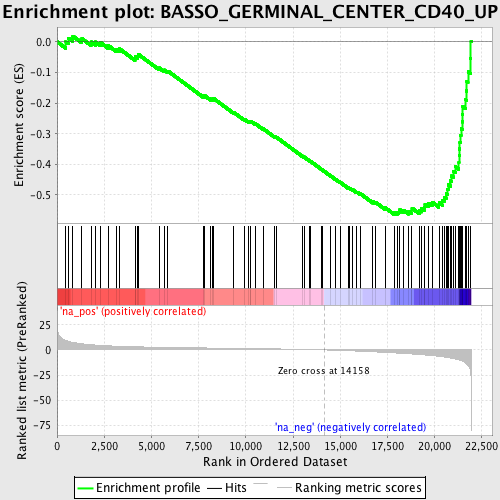
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BASSO_GERMINAL_CENTER_CD40_UP |
| Enrichment Score (ES) | -0.56361717 |
| Normalized Enrichment Score (NES) | -2.1386876 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0013714542 |
| FWER p-Value | 0.018 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UNC119 | 464 | 9.268 | -0.0003 | No | ||
| 2 | TNFAIP8 | 612 | 8.367 | 0.0119 | No | ||
| 3 | SYNGR2 | 834 | 7.454 | 0.0187 | No | ||
| 4 | HSP90B1 | 1314 | 6.000 | 0.0103 | No | ||
| 5 | TNFAIP2 | 1799 | 5.077 | -0.0004 | No | ||
| 6 | IER3 | 2035 | 4.764 | -0.0003 | No | ||
| 7 | PRG1 | 2305 | 4.414 | -0.0027 | No | ||
| 8 | MAPKAPK2 | 2720 | 3.998 | -0.0126 | No | ||
| 9 | CCL3 | 3147 | 3.696 | -0.0237 | No | ||
| 10 | TRIP10 | 3307 | 3.572 | -0.0229 | No | ||
| 11 | RXRA | 4129 | 3.082 | -0.0535 | No | ||
| 12 | PHACTR1 | 4150 | 3.069 | -0.0475 | No | ||
| 13 | LHFPL2 | 4284 | 3.002 | -0.0468 | No | ||
| 14 | LGALS1 | 4293 | 2.998 | -0.0403 | No | ||
| 15 | MEF2C | 5401 | 2.593 | -0.0851 | No | ||
| 16 | NCF2 | 5673 | 2.503 | -0.0919 | No | ||
| 17 | PLEK | 5872 | 2.439 | -0.0954 | No | ||
| 18 | ADAM8 | 7734 | 1.922 | -0.1762 | No | ||
| 19 | ICOSLG | 7828 | 1.900 | -0.1761 | No | ||
| 20 | IRF4 | 8141 | 1.829 | -0.1863 | No | ||
| 21 | CD44 | 8241 | 1.803 | -0.1867 | No | ||
| 22 | NEF3 | 8286 | 1.793 | -0.1847 | No | ||
| 23 | EHD1 | 9345 | 1.542 | -0.2296 | No | ||
| 24 | HTR3A | 9912 | 1.408 | -0.2523 | No | ||
| 25 | IFI30 | 10142 | 1.349 | -0.2597 | No | ||
| 26 | NFKBIE | 10229 | 1.326 | -0.2607 | No | ||
| 27 | LITAF | 10262 | 1.320 | -0.2592 | No | ||
| 28 | CCL4 | 10495 | 1.263 | -0.2669 | No | ||
| 29 | BLR1 | 10918 | 1.155 | -0.2836 | No | ||
| 30 | TCFL5 | 11544 | 0.981 | -0.3100 | No | ||
| 31 | PTP4A3 | 11613 | 0.963 | -0.3109 | No | ||
| 32 | CR2 | 12989 | 0.516 | -0.3726 | No | ||
| 33 | IRF5 | 13084 | 0.480 | -0.3759 | No | ||
| 34 | SNF1LK | 13371 | 0.369 | -0.3881 | No | ||
| 35 | FAIM3 | 13443 | 0.340 | -0.3906 | No | ||
| 36 | RGS1 | 14005 | 0.079 | -0.4161 | No | ||
| 37 | DDIT4 | 14068 | 0.043 | -0.4188 | No | ||
| 38 | CAPG | 14501 | -0.221 | -0.4381 | No | ||
| 39 | BATF | 14761 | -0.391 | -0.4490 | No | ||
| 40 | ELL2 | 15008 | -0.539 | -0.4591 | No | ||
| 41 | CD40 | 15416 | -0.823 | -0.4758 | No | ||
| 42 | CD74 | 15503 | -0.884 | -0.4778 | No | ||
| 43 | IL1B | 15675 | -1.000 | -0.4833 | No | ||
| 44 | PLXNC1 | 15885 | -1.144 | -0.4903 | No | ||
| 45 | TNF | 16079 | -1.307 | -0.4962 | No | ||
| 46 | CD83 | 16737 | -1.869 | -0.5220 | No | ||
| 47 | RAC2 | 16856 | -1.992 | -0.5229 | No | ||
| 48 | ICAM1 | 17395 | -2.482 | -0.5419 | No | ||
| 49 | MAP3K8 | 17860 | -2.954 | -0.5564 | No | ||
| 50 | ARID5B | 18018 | -3.120 | -0.5566 | Yes | ||
| 51 | TMC6 | 18117 | -3.239 | -0.5537 | Yes | ||
| 52 | RAB27A | 18166 | -3.290 | -0.5485 | Yes | ||
| 53 | SH2B3 | 18356 | -3.484 | -0.5493 | Yes | ||
| 54 | TNFSF9 | 18641 | -3.795 | -0.5537 | Yes | ||
| 55 | NCKAP1L | 18803 | -4.005 | -0.5520 | Yes | ||
| 56 | TAP1 | 18804 | -4.005 | -0.5429 | Yes | ||
| 57 | SNN | 19215 | -4.604 | -0.5513 | Yes | ||
| 58 | MTMR4 | 19309 | -4.743 | -0.5448 | Yes | ||
| 59 | LTA | 19451 | -4.972 | -0.5400 | Yes | ||
| 60 | NFKB2 | 19492 | -5.020 | -0.5305 | Yes | ||
| 61 | STAT5A | 19672 | -5.285 | -0.5267 | Yes | ||
| 62 | ARHGEF2 | 19872 | -5.614 | -0.5232 | Yes | ||
| 63 | TMSB4X | 20236 | -6.359 | -0.5254 | Yes | ||
| 64 | STK10 | 20418 | -6.774 | -0.5183 | Yes | ||
| 65 | KCNN4 | 20512 | -6.982 | -0.5068 | Yes | ||
| 66 | CCL22 | 20615 | -7.260 | -0.4951 | Yes | ||
| 67 | MYB | 20674 | -7.418 | -0.4810 | Yes | ||
| 68 | CHD1 | 20726 | -7.565 | -0.4662 | Yes | ||
| 69 | DTX2 | 20849 | -7.975 | -0.4537 | Yes | ||
| 70 | PIK3CD | 20906 | -8.191 | -0.4378 | Yes | ||
| 71 | TRAF1 | 20992 | -8.556 | -0.4223 | Yes | ||
| 72 | DUSP2 | 21113 | -8.980 | -0.4075 | Yes | ||
| 73 | ZFP36L1 | 21292 | -9.857 | -0.3934 | Yes | ||
| 74 | JUNB | 21298 | -9.897 | -0.3712 | Yes | ||
| 75 | ISGF3G | 21317 | -9.981 | -0.3495 | Yes | ||
| 76 | TCEB3 | 21344 | -10.143 | -0.3278 | Yes | ||
| 77 | BCL2 | 21371 | -10.300 | -0.3057 | Yes | ||
| 78 | GADD45B | 21406 | -10.572 | -0.2833 | Yes | ||
| 79 | CFLAR | 21465 | -10.989 | -0.2611 | Yes | ||
| 80 | TNFAIP3 | 21482 | -11.148 | -0.2367 | Yes | ||
| 81 | EBI2 | 21497 | -11.298 | -0.2118 | Yes | ||
| 82 | STAT1 | 21656 | -13.091 | -0.1894 | Yes | ||
| 83 | PTK2B | 21693 | -13.678 | -0.1601 | Yes | ||
| 84 | LAPTM5 | 21712 | -14.002 | -0.1293 | Yes | ||
| 85 | NFKBIA | 21775 | -15.114 | -0.0980 | Yes | ||
| 86 | SLAMF1 | 21917 | -21.771 | -0.0552 | Yes | ||
| 87 | CCR7 | 21931 | -25.001 | 0.0007 | Yes |
