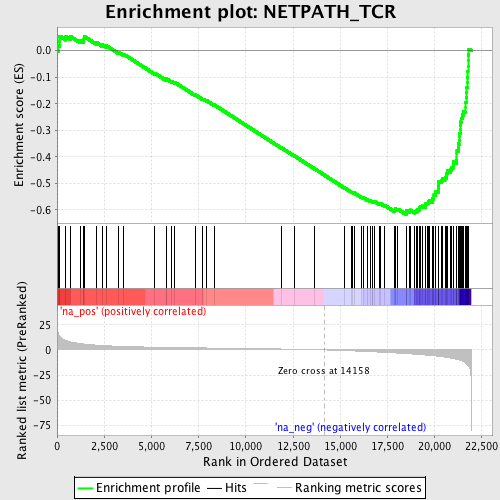
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | -0.6171192 |
| Normalized Enrichment Score (NES) | -2.4050918 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 74 | 15.638 | 0.0189 | No | ||
| 2 | NCL | 125 | 13.890 | 0.0364 | No | ||
| 3 | YWHAQ | 140 | 13.364 | 0.0548 | No | ||
| 4 | UNC119 | 464 | 9.268 | 0.0532 | No | ||
| 5 | CRK | 726 | 7.882 | 0.0524 | No | ||
| 6 | SHB | 1242 | 6.175 | 0.0376 | No | ||
| 7 | ACP1 | 1404 | 5.803 | 0.0385 | No | ||
| 8 | MAPK1 | 1423 | 5.768 | 0.0459 | No | ||
| 9 | PIK3R2 | 1448 | 5.700 | 0.0529 | No | ||
| 10 | DTX1 | 2076 | 4.725 | 0.0310 | No | ||
| 11 | NFATC2 | 2387 | 4.322 | 0.0229 | No | ||
| 12 | TUBB2A | 2614 | 4.097 | 0.0184 | No | ||
| 13 | ENAH | 3272 | 3.606 | -0.0066 | No | ||
| 14 | DUSP3 | 3516 | 3.427 | -0.0128 | No | ||
| 15 | STK39 | 5170 | 2.662 | -0.0847 | No | ||
| 16 | PTPRJ | 5775 | 2.469 | -0.1088 | No | ||
| 17 | SOS1 | 5789 | 2.466 | -0.1059 | No | ||
| 18 | NFAM1 | 6072 | 2.374 | -0.1155 | No | ||
| 19 | PIK3R1 | 6230 | 2.327 | -0.1193 | No | ||
| 20 | CDC42 | 7329 | 2.030 | -0.1667 | No | ||
| 21 | GAB2 | 7726 | 1.924 | -0.1821 | No | ||
| 22 | PTPN3 | 7927 | 1.877 | -0.1886 | No | ||
| 23 | VAV3 | 8314 | 1.784 | -0.2037 | No | ||
| 24 | HOMER3 | 11912 | 0.867 | -0.3672 | No | ||
| 25 | JUN | 12565 | 0.666 | -0.3961 | No | ||
| 26 | RAP1A | 13631 | 0.258 | -0.4445 | No | ||
| 27 | SRC | 15241 | -0.705 | -0.5172 | No | ||
| 28 | GRB2 | 15610 | -0.952 | -0.5327 | No | ||
| 29 | LAT | 15646 | -0.971 | -0.5329 | No | ||
| 30 | MAP2K1 | 15741 | -1.040 | -0.5357 | No | ||
| 31 | RIPK2 | 16143 | -1.360 | -0.5521 | No | ||
| 32 | CD2AP | 16239 | -1.438 | -0.5545 | No | ||
| 33 | ZAP70 | 16444 | -1.608 | -0.5615 | No | ||
| 34 | SOS2 | 16589 | -1.746 | -0.5656 | No | ||
| 35 | CTNNB1 | 16702 | -1.841 | -0.5681 | No | ||
| 36 | SHC1 | 16723 | -1.857 | -0.5664 | No | ||
| 37 | LIME1 | 16821 | -1.947 | -0.5681 | No | ||
| 38 | AKT1 | 17066 | -2.193 | -0.5761 | No | ||
| 39 | NCK1 | 17148 | -2.262 | -0.5766 | No | ||
| 40 | PTPN6 | 17353 | -2.437 | -0.5825 | No | ||
| 41 | PTPN22 | 17874 | -2.968 | -0.6021 | No | ||
| 42 | ARHGEF6 | 17905 | -3.006 | -0.5991 | No | ||
| 43 | JAK3 | 17929 | -3.030 | -0.5959 | No | ||
| 44 | SH2D2A | 18056 | -3.159 | -0.5972 | No | ||
| 45 | SYK | 18493 | -3.653 | -0.6119 | Yes | ||
| 46 | PAG1 | 18498 | -3.656 | -0.6069 | Yes | ||
| 47 | DBNL | 18517 | -3.675 | -0.6025 | Yes | ||
| 48 | FYB | 18658 | -3.819 | -0.6035 | Yes | ||
| 49 | PTPN11 | 18707 | -3.890 | -0.6001 | Yes | ||
| 50 | CREB1 | 18962 | -4.227 | -0.6057 | Yes | ||
| 51 | PTPN12 | 19048 | -4.339 | -0.6034 | Yes | ||
| 52 | KHDRBS1 | 19091 | -4.406 | -0.5991 | Yes | ||
| 53 | FOS | 19178 | -4.546 | -0.5966 | Yes | ||
| 54 | WAS | 19208 | -4.589 | -0.5914 | Yes | ||
| 55 | CD5 | 19230 | -4.624 | -0.5857 | Yes | ||
| 56 | CD3G | 19364 | -4.837 | -0.5849 | Yes | ||
| 57 | SH2D3C | 19505 | -5.042 | -0.5842 | Yes | ||
| 58 | PTPRC | 19517 | -5.062 | -0.5775 | Yes | ||
| 59 | CBLB | 19603 | -5.196 | -0.5740 | Yes | ||
| 60 | STAT5A | 19672 | -5.285 | -0.5695 | Yes | ||
| 61 | ABL1 | 19742 | -5.391 | -0.5650 | Yes | ||
| 62 | SH3BP2 | 19894 | -5.655 | -0.5639 | Yes | ||
| 63 | LAX1 | 19906 | -5.674 | -0.5563 | Yes | ||
| 64 | GRAP2 | 19967 | -5.818 | -0.5508 | Yes | ||
| 65 | CD3D | 19969 | -5.819 | -0.5425 | Yes | ||
| 66 | VAV1 | 20066 | -6.018 | -0.5384 | Yes | ||
| 67 | SKAP2 | 20068 | -6.019 | -0.5298 | Yes | ||
| 68 | TRAT1 | 20193 | -6.255 | -0.5266 | Yes | ||
| 69 | PLCG1 | 20195 | -6.259 | -0.5178 | Yes | ||
| 70 | DEF6 | 20211 | -6.300 | -0.5095 | Yes | ||
| 71 | DNM2 | 20229 | -6.348 | -0.5012 | Yes | ||
| 72 | BCL10 | 20231 | -6.352 | -0.4922 | Yes | ||
| 73 | TXK | 20361 | -6.619 | -0.4887 | Yes | ||
| 74 | ARHGEF7 | 20413 | -6.769 | -0.4814 | Yes | ||
| 75 | CRKL | 20555 | -7.087 | -0.4778 | Yes | ||
| 76 | SLA2 | 20618 | -7.270 | -0.4702 | Yes | ||
| 77 | PTK2 | 20652 | -7.365 | -0.4613 | Yes | ||
| 78 | DLG1 | 20702 | -7.506 | -0.4528 | Yes | ||
| 79 | RASA1 | 20833 | -7.927 | -0.4475 | Yes | ||
| 80 | PRKCQ | 20917 | -8.226 | -0.4396 | Yes | ||
| 81 | CISH | 20981 | -8.501 | -0.4304 | Yes | ||
| 82 | ABI1 | 21003 | -8.586 | -0.4191 | Yes | ||
| 83 | RAPGEF1 | 21148 | -9.136 | -0.4127 | Yes | ||
| 84 | CD8A | 21185 | -9.290 | -0.4011 | Yes | ||
| 85 | CARD11 | 21186 | -9.295 | -0.3879 | Yes | ||
| 86 | CD4 | 21188 | -9.300 | -0.3747 | Yes | ||
| 87 | EVL | 21269 | -9.694 | -0.3645 | Yes | ||
| 88 | SLA | 21278 | -9.796 | -0.3510 | Yes | ||
| 89 | FYN | 21301 | -9.911 | -0.3378 | Yes | ||
| 90 | ITK | 21334 | -10.083 | -0.3250 | Yes | ||
| 91 | ARHGDIB | 21340 | -10.120 | -0.3108 | Yes | ||
| 92 | DOCK2 | 21381 | -10.369 | -0.2978 | Yes | ||
| 93 | PSTPIP1 | 21385 | -10.420 | -0.2831 | Yes | ||
| 94 | GRAP | 21394 | -10.473 | -0.2686 | Yes | ||
| 95 | CREBBP | 21411 | -10.605 | -0.2542 | Yes | ||
| 96 | WASF2 | 21477 | -11.047 | -0.2415 | Yes | ||
| 97 | CD3E | 21545 | -11.647 | -0.2280 | Yes | ||
| 98 | STAT5B | 21655 | -13.076 | -0.2143 | Yes | ||
| 99 | GIT2 | 21657 | -13.105 | -0.1957 | Yes | ||
| 100 | PRKD2 | 21683 | -13.494 | -0.1777 | Yes | ||
| 101 | PTK2B | 21693 | -13.678 | -0.1586 | Yes | ||
| 102 | CBL | 21707 | -13.898 | -0.1394 | Yes | ||
| 103 | SIT1 | 21731 | -14.291 | -0.1201 | Yes | ||
| 104 | VASP | 21737 | -14.401 | -0.0998 | Yes | ||
| 105 | LCP2 | 21764 | -14.815 | -0.0799 | Yes | ||
| 106 | SKAP1 | 21774 | -15.080 | -0.0589 | Yes | ||
| 107 | LCK | 21778 | -15.166 | -0.0374 | Yes | ||
| 108 | CD247 | 21783 | -15.293 | -0.0158 | Yes | ||
| 109 | NEDD9 | 21820 | -16.353 | 0.0058 | Yes |