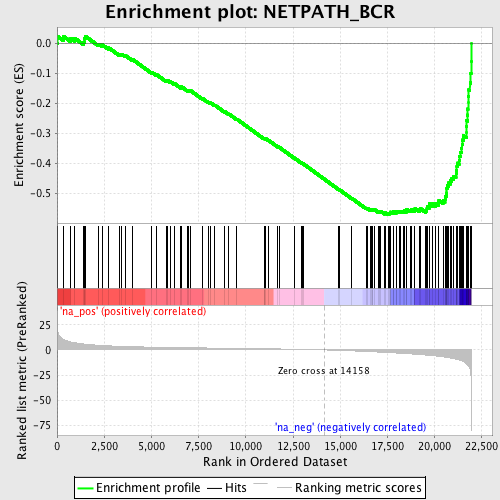
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.5694712 |
| Normalized Enrichment Score (NES) | -2.2988634 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CYCS | 42 | 17.128 | 0.0222 | No | ||
| 2 | CDK4 | 343 | 10.371 | 0.0231 | No | ||
| 3 | CRK | 726 | 7.882 | 0.0167 | No | ||
| 4 | FCGR2B | 927 | 7.120 | 0.0176 | No | ||
| 5 | MAPK1 | 1423 | 5.768 | 0.0030 | No | ||
| 6 | PIK3R2 | 1448 | 5.700 | 0.0100 | No | ||
| 7 | BLK | 1450 | 5.692 | 0.0179 | No | ||
| 8 | CCNE1 | 1485 | 5.615 | 0.0243 | No | ||
| 9 | BAX | 2201 | 4.543 | -0.0020 | No | ||
| 10 | NFATC2 | 2387 | 4.322 | -0.0044 | No | ||
| 11 | MAPKAPK2 | 2720 | 3.998 | -0.0140 | No | ||
| 12 | CCNA2 | 3305 | 3.575 | -0.0357 | No | ||
| 13 | CASP9 | 3432 | 3.473 | -0.0366 | No | ||
| 14 | ITPR2 | 3601 | 3.360 | -0.0396 | No | ||
| 15 | BTK | 3984 | 3.157 | -0.0526 | No | ||
| 16 | DOK3 | 5026 | 2.710 | -0.0965 | No | ||
| 17 | RPS6KA1 | 5264 | 2.635 | -0.1036 | No | ||
| 18 | SOS1 | 5789 | 2.466 | -0.1242 | No | ||
| 19 | ATP2B4 | 5841 | 2.448 | -0.1230 | No | ||
| 20 | PIK3AP1 | 5991 | 2.399 | -0.1265 | No | ||
| 21 | PIK3R1 | 6230 | 2.327 | -0.1341 | No | ||
| 22 | MAPK4 | 6559 | 2.234 | -0.1460 | No | ||
| 23 | HCK | 6566 | 2.233 | -0.1431 | No | ||
| 24 | LAT2 | 6919 | 2.136 | -0.1562 | No | ||
| 25 | MAP3K7 | 6975 | 2.124 | -0.1558 | No | ||
| 26 | CD72 | 7056 | 2.104 | -0.1565 | No | ||
| 27 | GAB2 | 7726 | 1.924 | -0.1844 | No | ||
| 28 | CD19 | 8029 | 1.852 | -0.1956 | No | ||
| 29 | CDK6 | 8138 | 1.830 | -0.1980 | No | ||
| 30 | CD22 | 8363 | 1.778 | -0.2057 | No | ||
| 31 | CD79A | 8876 | 1.654 | -0.2269 | No | ||
| 32 | PPP3CA | 9099 | 1.601 | -0.2348 | No | ||
| 33 | PRKD1 | 9525 | 1.498 | -0.2521 | No | ||
| 34 | RASGRP3 | 10969 | 1.145 | -0.3166 | No | ||
| 35 | CDK2 | 10991 | 1.139 | -0.3160 | No | ||
| 36 | BCAR1 | 11044 | 1.123 | -0.3168 | No | ||
| 37 | RPS6 | 11187 | 1.089 | -0.3218 | No | ||
| 38 | BLNK | 11697 | 0.938 | -0.3438 | No | ||
| 39 | DOK1 | 11766 | 0.915 | -0.3456 | No | ||
| 40 | JUN | 12565 | 0.666 | -0.3812 | No | ||
| 41 | CD81 | 12940 | 0.537 | -0.3976 | No | ||
| 42 | CR2 | 12989 | 0.516 | -0.3991 | No | ||
| 43 | PIP5K1C | 13033 | 0.503 | -0.4003 | No | ||
| 44 | REL | 14913 | -0.486 | -0.4857 | No | ||
| 45 | GSK3A | 14949 | -0.501 | -0.4866 | No | ||
| 46 | GRB2 | 15610 | -0.952 | -0.5155 | No | ||
| 47 | GTF2I | 16370 | -1.548 | -0.5481 | No | ||
| 48 | ZAP70 | 16444 | -1.608 | -0.5492 | No | ||
| 49 | SOS2 | 16589 | -1.746 | -0.5533 | No | ||
| 50 | ACTR3 | 16633 | -1.785 | -0.5528 | No | ||
| 51 | CTNNB1 | 16702 | -1.841 | -0.5533 | No | ||
| 52 | SHC1 | 16723 | -1.857 | -0.5516 | No | ||
| 53 | LIME1 | 16821 | -1.947 | -0.5533 | No | ||
| 54 | HCLS1 | 17013 | -2.142 | -0.5590 | No | ||
| 55 | AKT1 | 17066 | -2.193 | -0.5583 | No | ||
| 56 | NCK1 | 17148 | -2.262 | -0.5588 | No | ||
| 57 | PTPN6 | 17353 | -2.437 | -0.5647 | No | ||
| 58 | RAP2A | 17415 | -2.498 | -0.5640 | No | ||
| 59 | PLEKHA1 | 17535 | -2.637 | -0.5658 | Yes | ||
| 60 | GAB1 | 17593 | -2.683 | -0.5646 | Yes | ||
| 61 | CHUK | 17643 | -2.730 | -0.5630 | Yes | ||
| 62 | ELK1 | 17690 | -2.773 | -0.5612 | Yes | ||
| 63 | MAPK14 | 17811 | -2.903 | -0.5626 | Yes | ||
| 64 | HDAC5 | 17835 | -2.933 | -0.5595 | Yes | ||
| 65 | MAPK3 | 17962 | -3.067 | -0.5609 | Yes | ||
| 66 | CDK7 | 18002 | -3.105 | -0.5584 | Yes | ||
| 67 | PTPN18 | 18127 | -3.246 | -0.5595 | Yes | ||
| 68 | PPP3R1 | 18216 | -3.345 | -0.5588 | Yes | ||
| 69 | BRAF | 18334 | -3.456 | -0.5593 | Yes | ||
| 70 | IKBKG | 18397 | -3.530 | -0.5571 | Yes | ||
| 71 | SYK | 18493 | -3.653 | -0.5563 | Yes | ||
| 72 | PRKCD | 18528 | -3.688 | -0.5527 | Yes | ||
| 73 | PTPN11 | 18707 | -3.890 | -0.5554 | Yes | ||
| 74 | RB1 | 18763 | -3.958 | -0.5523 | Yes | ||
| 75 | DAPP1 | 18923 | -4.178 | -0.5537 | Yes | ||
| 76 | CREB1 | 18962 | -4.227 | -0.5495 | Yes | ||
| 77 | WAS | 19208 | -4.589 | -0.5542 | Yes | ||
| 78 | CD5 | 19230 | -4.624 | -0.5487 | Yes | ||
| 79 | PTPRC | 19517 | -5.062 | -0.5546 | Yes | ||
| 80 | CD79B | 19598 | -5.186 | -0.5510 | Yes | ||
| 81 | CBLB | 19603 | -5.196 | -0.5439 | Yes | ||
| 82 | ATF2 | 19713 | -5.352 | -0.5413 | Yes | ||
| 83 | IKBKB | 19718 | -5.359 | -0.5339 | Yes | ||
| 84 | SH3BP2 | 19894 | -5.655 | -0.5340 | Yes | ||
| 85 | VAV1 | 20066 | -6.018 | -0.5333 | Yes | ||
| 86 | PLCG1 | 20195 | -6.259 | -0.5304 | Yes | ||
| 87 | BCL10 | 20231 | -6.352 | -0.5230 | Yes | ||
| 88 | VAV2 | 20452 | -6.862 | -0.5234 | Yes | ||
| 89 | CRKL | 20555 | -7.087 | -0.5181 | Yes | ||
| 90 | BCL2L11 | 20565 | -7.114 | -0.5085 | Yes | ||
| 91 | SLA2 | 20618 | -7.270 | -0.5006 | Yes | ||
| 92 | PTK2 | 20652 | -7.365 | -0.4918 | Yes | ||
| 93 | PRKCE | 20653 | -7.365 | -0.4814 | Yes | ||
| 94 | ACTR2 | 20688 | -7.465 | -0.4724 | Yes | ||
| 95 | PDK2 | 20750 | -7.630 | -0.4644 | Yes | ||
| 96 | RASA1 | 20833 | -7.927 | -0.4570 | Yes | ||
| 97 | PRKCQ | 20917 | -8.226 | -0.4492 | Yes | ||
| 98 | RELA | 21022 | -8.671 | -0.4418 | Yes | ||
| 99 | CCND2 | 21139 | -9.105 | -0.4343 | Yes | ||
| 100 | RAPGEF1 | 21148 | -9.136 | -0.4217 | Yes | ||
| 101 | CARD11 | 21186 | -9.295 | -0.4103 | Yes | ||
| 102 | MAPK8 | 21208 | -9.408 | -0.3980 | Yes | ||
| 103 | FYN | 21301 | -9.911 | -0.3883 | Yes | ||
| 104 | ITK | 21334 | -10.083 | -0.3755 | Yes | ||
| 105 | BCL2 | 21371 | -10.300 | -0.3627 | Yes | ||
| 106 | CSK | 21416 | -10.654 | -0.3497 | Yes | ||
| 107 | GSK3B | 21455 | -10.935 | -0.3360 | Yes | ||
| 108 | RPS6KB1 | 21479 | -11.098 | -0.3214 | Yes | ||
| 109 | NFATC1 | 21534 | -11.557 | -0.3076 | Yes | ||
| 110 | PTK2B | 21693 | -13.678 | -0.2955 | Yes | ||
| 111 | TEC | 21697 | -13.757 | -0.2763 | Yes | ||
| 112 | CBL | 21707 | -13.898 | -0.2571 | Yes | ||
| 113 | NFATC3 | 21743 | -14.538 | -0.2382 | Yes | ||
| 114 | LCP2 | 21764 | -14.815 | -0.2182 | Yes | ||
| 115 | NFKBIA | 21775 | -15.114 | -0.1974 | Yes | ||
| 116 | LCK | 21778 | -15.166 | -0.1761 | Yes | ||
| 117 | NEDD9 | 21820 | -16.353 | -0.1549 | Yes | ||
| 118 | PDPK1 | 21879 | -18.696 | -0.1312 | Yes | ||
| 119 | CCND3 | 21924 | -22.833 | -0.1011 | Yes | ||
| 120 | CASP7 | 21938 | -29.718 | -0.0598 | Yes | ||
| 121 | BCL6 | 21944 | -42.648 | 0.0001 | Yes |