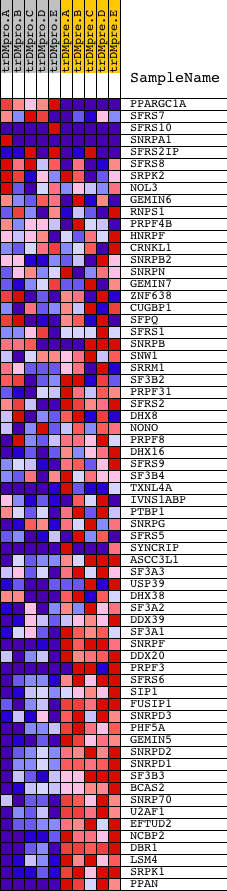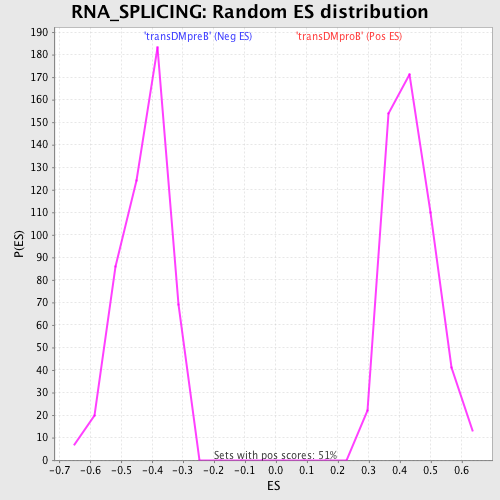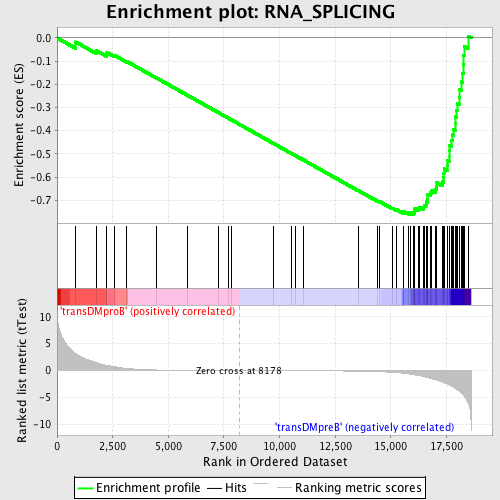
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | RNA_SPLICING |
| Enrichment Score (ES) | -0.7613393 |
| Normalized Enrichment Score (NES) | -1.791115 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0015931266 |
| FWER p-Value | 0.0070 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPARGC1A | 16533 | 835 | 3.159 | -0.0175 | No | ||
| 2 | SFRS7 | 22889 | 1763 | 1.472 | -0.0546 | No | ||
| 3 | SFRS10 | 9818 1722 5441 | 2217 | 0.998 | -0.0704 | No | ||
| 4 | SNRPA1 | 1373 18216 | 2234 | 0.991 | -0.0626 | No | ||
| 5 | SFRS2IP | 7794 13009 | 2590 | 0.706 | -0.0756 | No | ||
| 6 | SFRS8 | 10543 6089 | 3124 | 0.394 | -0.1009 | No | ||
| 7 | SRPK2 | 5513 | 4466 | 0.095 | -0.1723 | No | ||
| 8 | NOL3 | 18496 915 921 | 5871 | 0.037 | -0.2477 | No | ||
| 9 | GEMIN6 | 12492 7415 | 7235 | 0.013 | -0.3211 | No | ||
| 10 | RNPS1 | 9730 23361 | 7695 | 0.007 | -0.3457 | No | ||
| 11 | PRPF4B | 3208 5295 | 7854 | 0.004 | -0.3542 | No | ||
| 12 | HNRPF | 13607 | 9743 | -0.020 | -0.4558 | No | ||
| 13 | CRNKL1 | 14407 | 10520 | -0.031 | -0.4974 | No | ||
| 14 | SNRPB2 | 5470 | 10695 | -0.034 | -0.5065 | No | ||
| 15 | SNRPN | 1715 9844 5471 | 11092 | -0.041 | -0.5275 | No | ||
| 16 | GEMIN7 | 17944 | 13533 | -0.124 | -0.6579 | No | ||
| 17 | ZNF638 | 9476 | 14406 | -0.211 | -0.7031 | No | ||
| 18 | CUGBP1 | 2805 8819 4576 2924 | 14499 | -0.225 | -0.7061 | No | ||
| 19 | SFPQ | 12936 | 15076 | -0.341 | -0.7342 | No | ||
| 20 | SFRS1 | 8492 | 15244 | -0.391 | -0.7398 | No | ||
| 21 | SNRPB | 9842 5469 2736 | 15571 | -0.515 | -0.7528 | No | ||
| 22 | SNW1 | 7282 | 15575 | -0.516 | -0.7485 | No | ||
| 23 | SRRM1 | 6958 | 15782 | -0.618 | -0.7542 | No | ||
| 24 | SF3B2 | 23974 | 15888 | -0.682 | -0.7540 | No | ||
| 25 | PRPF31 | 7594 | 16026 | -0.782 | -0.7545 | Yes | ||
| 26 | SFRS2 | 9807 20136 | 16049 | -0.798 | -0.7488 | Yes | ||
| 27 | DHX8 | 20649 | 16062 | -0.808 | -0.7424 | Yes | ||
| 28 | NONO | 11993 | 16084 | -0.831 | -0.7363 | Yes | ||
| 29 | PRPF8 | 20780 1371 | 16257 | -0.972 | -0.7371 | Yes | ||
| 30 | DHX16 | 23247 | 16271 | -0.980 | -0.7292 | Yes | ||
| 31 | SFRS9 | 16731 | 16466 | -1.109 | -0.7301 | Yes | ||
| 32 | SF3B4 | 22269 | 16492 | -1.131 | -0.7215 | Yes | ||
| 33 | TXNL4A | 6567 11329 | 16582 | -1.228 | -0.7157 | Yes | ||
| 34 | IVNS1ABP | 4384 4050 | 16604 | -1.245 | -0.7059 | Yes | ||
| 35 | PTBP1 | 5303 | 16652 | -1.298 | -0.6972 | Yes | ||
| 36 | SNRPG | 12622 | 16664 | -1.317 | -0.6863 | Yes | ||
| 37 | SFRS5 | 9808 2062 | 16665 | -1.319 | -0.6748 | Yes | ||
| 38 | SYNCRIP | 3078 3035 3107 | 16783 | -1.471 | -0.6683 | Yes | ||
| 39 | ASCC3L1 | 14867 | 16848 | -1.547 | -0.6583 | Yes | ||
| 40 | SF3A3 | 16091 | 17000 | -1.699 | -0.6516 | Yes | ||
| 41 | USP39 | 1116 1083 11373 | 17047 | -1.748 | -0.6389 | Yes | ||
| 42 | DHX38 | 48 | 17069 | -1.776 | -0.6245 | Yes | ||
| 43 | SF3A2 | 19938 | 17326 | -2.138 | -0.6197 | Yes | ||
| 44 | DDX39 | 18551 | 17362 | -2.193 | -0.6025 | Yes | ||
| 45 | SF3A1 | 7450 | 17384 | -2.247 | -0.5841 | Yes | ||
| 46 | SNRPF | 7645 | 17393 | -2.259 | -0.5648 | Yes | ||
| 47 | DDX20 | 15213 | 17544 | -2.502 | -0.5511 | Yes | ||
| 48 | PRPF3 | 12899 12898 1877 7703 | 17562 | -2.549 | -0.5298 | Yes | ||
| 49 | SFRS6 | 14751 | 17617 | -2.689 | -0.5093 | Yes | ||
| 50 | SIP1 | 21263 | 17619 | -2.696 | -0.4859 | Yes | ||
| 51 | FUSIP1 | 4715 16036 | 17627 | -2.708 | -0.4627 | Yes | ||
| 52 | SNRPD3 | 12514 | 17723 | -2.911 | -0.4424 | Yes | ||
| 53 | PHF5A | 22194 | 17755 | -2.970 | -0.4182 | Yes | ||
| 54 | GEMIN5 | 20439 | 17808 | -3.106 | -0.3940 | Yes | ||
| 55 | SNRPD2 | 8412 | 17892 | -3.333 | -0.3694 | Yes | ||
| 56 | SNRPD1 | 23622 | 17897 | -3.342 | -0.3405 | Yes | ||
| 57 | SF3B3 | 18746 | 17955 | -3.510 | -0.3131 | Yes | ||
| 58 | BCAS2 | 12662 | 17977 | -3.573 | -0.2831 | Yes | ||
| 59 | SNRP70 | 1186 | 18077 | -3.843 | -0.2549 | Yes | ||
| 60 | U2AF1 | 8431 | 18097 | -3.895 | -0.2220 | Yes | ||
| 61 | EFTUD2 | 1219 20203 | 18188 | -4.255 | -0.1898 | Yes | ||
| 62 | NCBP2 | 12643 | 18234 | -4.445 | -0.1535 | Yes | ||
| 63 | DBR1 | 19341 | 18259 | -4.531 | -0.1154 | Yes | ||
| 64 | LSM4 | 18583 | 18282 | -4.650 | -0.0760 | Yes | ||
| 65 | SRPK1 | 23049 | 18311 | -4.799 | -0.0358 | Yes | ||
| 66 | PPAN | 19546 | 18470 | -5.987 | 0.0079 | Yes |