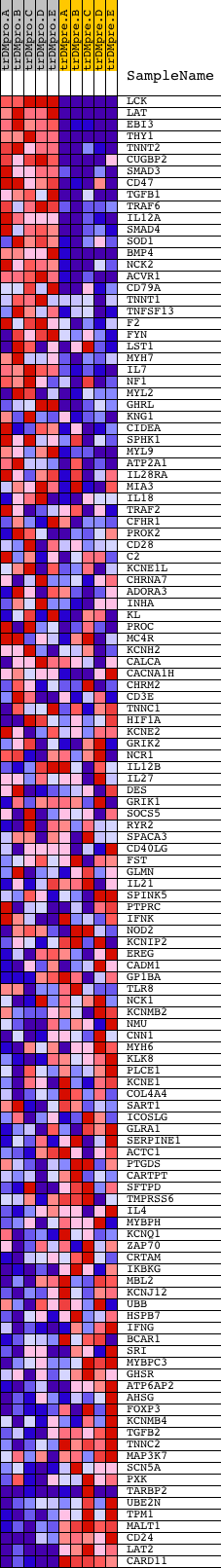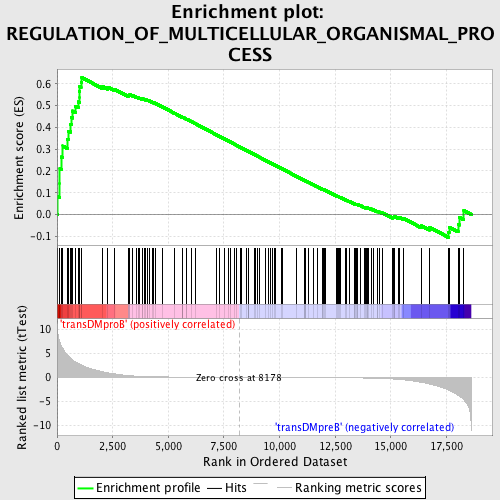
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | REGULATION_OF_MULTICELLULAR_ORGANISMAL_PROCESS |
| Enrichment Score (ES) | 0.6283833 |
| Normalized Enrichment Score (NES) | 1.5816913 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.26843473 |
| FWER p-Value | 0.815 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LCK | 15746 | 27 | 9.283 | 0.0828 | Yes | ||
| 2 | LAT | 17643 | 124 | 7.317 | 0.1441 | Yes | ||
| 3 | EBI3 | 23193 | 127 | 7.299 | 0.2103 | Yes | ||
| 4 | THY1 | 19481 | 199 | 6.383 | 0.2644 | Yes | ||
| 5 | TNNT2 | 14113 | 262 | 5.908 | 0.3147 | Yes | ||
| 6 | CUGBP2 | 4687 8923 | 457 | 4.622 | 0.3462 | Yes | ||
| 7 | SMAD3 | 19084 | 511 | 4.367 | 0.3830 | Yes | ||
| 8 | CD47 | 4933 | 596 | 3.950 | 0.4143 | Yes | ||
| 9 | TGFB1 | 18332 | 647 | 3.751 | 0.4457 | Yes | ||
| 10 | TRAF6 | 5797 14940 | 701 | 3.574 | 0.4753 | Yes | ||
| 11 | IL12A | 4913 | 820 | 3.198 | 0.4979 | Yes | ||
| 12 | SMAD4 | 5058 | 952 | 2.856 | 0.5168 | Yes | ||
| 13 | SOD1 | 9846 | 1001 | 2.740 | 0.5391 | Yes | ||
| 14 | BMP4 | 21863 | 1004 | 2.727 | 0.5637 | Yes | ||
| 15 | NCK2 | 9448 | 1018 | 2.695 | 0.5875 | Yes | ||
| 16 | ACVR1 | 4334 | 1076 | 2.564 | 0.6077 | Yes | ||
| 17 | CD79A | 18342 | 1111 | 2.480 | 0.6284 | Yes | ||
| 18 | TNNT1 | 17988 | 2053 | 1.111 | 0.5876 | No | ||
| 19 | TNFSF13 | 12806 | 2276 | 0.962 | 0.5843 | No | ||
| 20 | F2 | 14524 | 2573 | 0.720 | 0.5748 | No | ||
| 21 | FYN | 3375 3395 20052 | 3206 | 0.359 | 0.5439 | No | ||
| 22 | LST1 | 9287 | 3228 | 0.348 | 0.5460 | No | ||
| 23 | MYH7 | 21828 4709 | 3242 | 0.343 | 0.5484 | No | ||
| 24 | IL7 | 4921 | 3269 | 0.334 | 0.5500 | No | ||
| 25 | NF1 | 5165 | 3387 | 0.293 | 0.5463 | No | ||
| 26 | MYL2 | 16713 | 3560 | 0.242 | 0.5392 | No | ||
| 27 | GHRL | 1183 17040 | 3675 | 0.216 | 0.5350 | No | ||
| 28 | KNG1 | 9244 22809 | 3724 | 0.205 | 0.5343 | No | ||
| 29 | CIDEA | 23529 | 3822 | 0.186 | 0.5307 | No | ||
| 30 | SPHK1 | 5484 1266 1402 | 3834 | 0.183 | 0.5318 | No | ||
| 31 | MYL9 | 14767 2659 | 3922 | 0.165 | 0.5286 | No | ||
| 32 | ATP2A1 | 17639 | 3986 | 0.155 | 0.5266 | No | ||
| 33 | IL28RA | 10823 16038 | 4059 | 0.143 | 0.5240 | No | ||
| 34 | MIA3 | 6868 6867 11679 | 4077 | 0.139 | 0.5243 | No | ||
| 35 | IL18 | 9172 | 4165 | 0.128 | 0.5208 | No | ||
| 36 | TRAF2 | 14657 | 4297 | 0.111 | 0.5147 | No | ||
| 37 | CFHR1 | 11926 13815 | 4354 | 0.105 | 0.5126 | No | ||
| 38 | PROK2 | 17057 | 4439 | 0.098 | 0.5090 | No | ||
| 39 | CD28 | 14239 4092 | 4745 | 0.076 | 0.4932 | No | ||
| 40 | C2 | 23013 | 5259 | 0.055 | 0.4659 | No | ||
| 41 | KCNE1L | 24044 | 5630 | 0.043 | 0.4463 | No | ||
| 42 | CHRNA7 | 17808 | 5632 | 0.043 | 0.4467 | No | ||
| 43 | ADORA3 | 4352 | 5653 | 0.043 | 0.4460 | No | ||
| 44 | INHA | 14214 | 5832 | 0.038 | 0.4367 | No | ||
| 45 | KL | 16611 | 5834 | 0.038 | 0.4370 | No | ||
| 46 | PROC | 23475 | 6022 | 0.034 | 0.4272 | No | ||
| 47 | MC4R | 13701 | 6234 | 0.029 | 0.4160 | No | ||
| 48 | KCNH2 | 16592 | 7166 | 0.014 | 0.3658 | No | ||
| 49 | CALCA | 4470 | 7316 | 0.012 | 0.3578 | No | ||
| 50 | CACNA1H | 23077 | 7538 | 0.009 | 0.3460 | No | ||
| 51 | CHRM2 | 10840 | 7691 | 0.007 | 0.3378 | No | ||
| 52 | CD3E | 8714 | 7771 | 0.006 | 0.3336 | No | ||
| 53 | TNNC1 | 22058 | 7965 | 0.003 | 0.3232 | No | ||
| 54 | HIF1A | 4850 | 8051 | 0.002 | 0.3186 | No | ||
| 55 | KCNE2 | 22701 | 8237 | -0.001 | 0.3086 | No | ||
| 56 | GRIK2 | 3318 | 8286 | -0.002 | 0.3060 | No | ||
| 57 | NCR1 | 18409 | 8522 | -0.004 | 0.2934 | No | ||
| 58 | IL12B | 20918 | 8591 | -0.005 | 0.2897 | No | ||
| 59 | IL27 | 17636 | 8592 | -0.005 | 0.2898 | No | ||
| 60 | DES | 14215 | 8604 | -0.005 | 0.2892 | No | ||
| 61 | GRIK1 | 22550 1741 | 8863 | -0.009 | 0.2754 | No | ||
| 62 | SOCS5 | 1619 23141 | 8896 | -0.009 | 0.2737 | No | ||
| 63 | RYR2 | 5403 | 9024 | -0.011 | 0.2669 | No | ||
| 64 | SPACA3 | 13680 | 9111 | -0.012 | 0.2624 | No | ||
| 65 | CD40LG | 24330 | 9346 | -0.015 | 0.2499 | No | ||
| 66 | FST | 8985 | 9511 | -0.017 | 0.2412 | No | ||
| 67 | GLMN | 16452 3562 | 9613 | -0.018 | 0.2359 | No | ||
| 68 | IL21 | 12241 | 9664 | -0.019 | 0.2333 | No | ||
| 69 | SPINK5 | 23569 | 9763 | -0.020 | 0.2282 | No | ||
| 70 | PTPRC | 5327 9662 | 9820 | -0.021 | 0.2254 | No | ||
| 71 | IFNK | 11837 | 10091 | -0.024 | 0.2110 | No | ||
| 72 | NOD2 | 6384 | 10120 | -0.025 | 0.2097 | No | ||
| 73 | KCNIP2 | 23655 3725 | 10757 | -0.035 | 0.1756 | No | ||
| 74 | EREG | 4679 16797 | 11104 | -0.041 | 0.1573 | No | ||
| 75 | CADM1 | 7057 | 11139 | -0.042 | 0.1558 | No | ||
| 76 | GP1BA | 13683 | 11180 | -0.042 | 0.1540 | No | ||
| 77 | TLR8 | 9308 | 11181 | -0.042 | 0.1544 | No | ||
| 78 | NCK1 | 9447 5152 | 11287 | -0.044 | 0.1491 | No | ||
| 79 | KCNMB2 | 1939 15615 | 11530 | -0.049 | 0.1365 | No | ||
| 80 | NMU | 16506 | 11708 | -0.053 | 0.1274 | No | ||
| 81 | CNN1 | 8759 | 11917 | -0.059 | 0.1167 | No | ||
| 82 | MYH6 | 9436 | 11940 | -0.059 | 0.1161 | No | ||
| 83 | KLK8 | 18269 18271 | 11979 | -0.060 | 0.1145 | No | ||
| 84 | PLCE1 | 13156 | 12001 | -0.061 | 0.1140 | No | ||
| 85 | KCNE1 | 9206 22537 4942 | 12084 | -0.063 | 0.1101 | No | ||
| 86 | COL4A4 | 8767 | 12557 | -0.078 | 0.0853 | No | ||
| 87 | SART1 | 23977 | 12589 | -0.079 | 0.0843 | No | ||
| 88 | ICOSLG | 19973 | 12628 | -0.081 | 0.0830 | No | ||
| 89 | GLRA1 | 9021 | 12691 | -0.083 | 0.0804 | No | ||
| 90 | SERPINE1 | 16333 | 12748 | -0.085 | 0.0781 | No | ||
| 91 | ACTC1 | 14481 14482 | 12958 | -0.093 | 0.0677 | No | ||
| 92 | PTGDS | 14658 | 12989 | -0.095 | 0.0669 | No | ||
| 93 | CARTPT | 21374 | 13154 | -0.103 | 0.0590 | No | ||
| 94 | SFTPD | 21867 | 13350 | -0.112 | 0.0495 | No | ||
| 95 | TMPRSS6 | 2263 22220 | 13374 | -0.114 | 0.0493 | No | ||
| 96 | IL4 | 9174 | 13421 | -0.117 | 0.0478 | No | ||
| 97 | MYBPH | 14130 | 13434 | -0.118 | 0.0482 | No | ||
| 98 | KCNQ1 | 18001 4948 | 13501 | -0.122 | 0.0458 | No | ||
| 99 | ZAP70 | 14271 4042 | 13513 | -0.123 | 0.0463 | No | ||
| 100 | CRTAM | 19160 | 13617 | -0.131 | 0.0419 | No | ||
| 101 | IKBKG | 2570 2562 4908 | 13814 | -0.147 | 0.0327 | No | ||
| 102 | MBL2 | 23886 | 13865 | -0.152 | 0.0313 | No | ||
| 103 | KCNJ12 | 20855 | 13897 | -0.155 | 0.0311 | No | ||
| 104 | UBB | 10240 | 13945 | -0.159 | 0.0300 | No | ||
| 105 | HSPB7 | 16005 | 13955 | -0.160 | 0.0309 | No | ||
| 106 | IFNG | 19869 | 14005 | -0.166 | 0.0298 | No | ||
| 107 | BCAR1 | 18741 | 14108 | -0.174 | 0.0259 | No | ||
| 108 | SRI | 3656 8462 | 14200 | -0.185 | 0.0226 | No | ||
| 109 | MYBPC3 | 9434 | 14399 | -0.210 | 0.0138 | No | ||
| 110 | GHSR | 1810 15625 | 14405 | -0.211 | 0.0155 | No | ||
| 111 | ATP6AP2 | 2567 24380 2579 2656 | 14512 | -0.226 | 0.0118 | No | ||
| 112 | AHSG | 22812 | 14630 | -0.245 | 0.0077 | No | ||
| 113 | FOXP3 | 9804 5426 | 15068 | -0.338 | -0.0129 | No | ||
| 114 | KCNMB4 | 12217 | 15114 | -0.353 | -0.0121 | No | ||
| 115 | TGFB2 | 5743 | 15170 | -0.369 | -0.0117 | No | ||
| 116 | TNNC2 | 14343 | 15176 | -0.371 | -0.0086 | No | ||
| 117 | MAP3K7 | 16255 | 15341 | -0.423 | -0.0137 | No | ||
| 118 | SCN5A | 5412 | 15375 | -0.432 | -0.0115 | No | ||
| 119 | PXK | 26 | 15581 | -0.518 | -0.0179 | No | ||
| 120 | TARBP2 | 10033 10034 5626 | 16381 | -1.030 | -0.0518 | No | ||
| 121 | UBE2N | 8216 19898 | 16734 | -1.407 | -0.0580 | No | ||
| 122 | TPM1 | 19070 | 17600 | -2.644 | -0.0808 | No | ||
| 123 | MALT1 | 6274 | 17621 | -2.701 | -0.0574 | No | ||
| 124 | CD24 | 8711 | 18026 | -3.725 | -0.0454 | No | ||
| 125 | LAT2 | 16351 | 18072 | -3.834 | -0.0130 | No | ||
| 126 | CARD11 | 8436 | 18284 | -4.663 | 0.0180 | No |