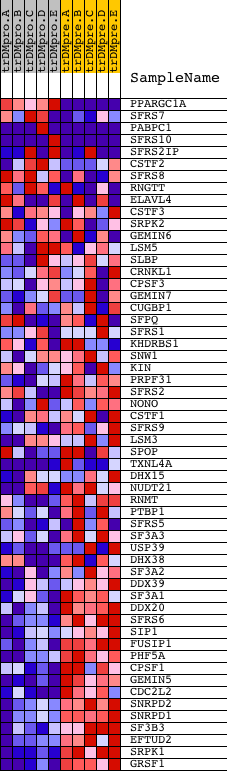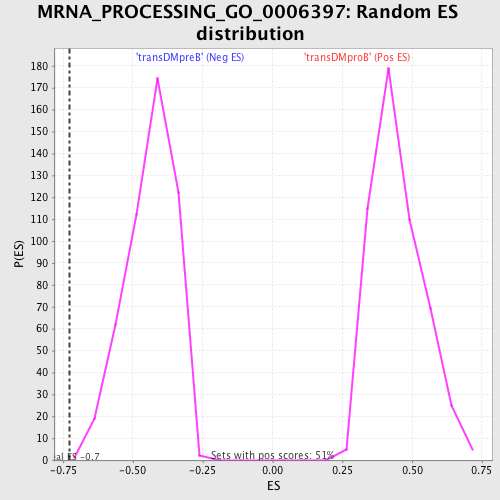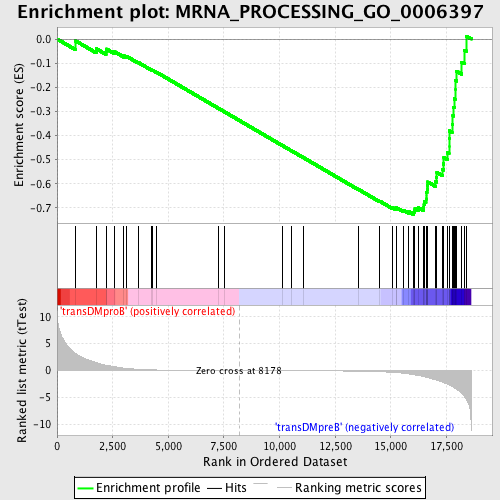
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | MRNA_PROCESSING_GO_0006397 |
| Enrichment Score (ES) | -0.7273973 |
| Normalized Enrichment Score (NES) | -1.6601306 |
| Nominal p-value | 0.0020325202 |
| FDR q-value | 0.029769436 |
| FWER p-Value | 0.307 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPARGC1A | 16533 | 835 | 3.159 | -0.0079 | No | ||
| 2 | SFRS7 | 22889 | 1763 | 1.472 | -0.0405 | No | ||
| 3 | PABPC1 | 5219 9522 9523 23572 | 2200 | 1.000 | -0.0522 | No | ||
| 4 | SFRS10 | 9818 1722 5441 | 2217 | 0.998 | -0.0414 | No | ||
| 5 | SFRS2IP | 7794 13009 | 2590 | 0.706 | -0.0531 | No | ||
| 6 | CSTF2 | 24257 | 2997 | 0.456 | -0.0696 | No | ||
| 7 | SFRS8 | 10543 6089 | 3124 | 0.394 | -0.0718 | No | ||
| 8 | RNGTT | 10769 2354 6272 | 3641 | 0.224 | -0.0969 | No | ||
| 9 | ELAVL4 | 15805 4889 9137 | 4250 | 0.116 | -0.1283 | No | ||
| 10 | CSTF3 | 10449 6002 | 4295 | 0.111 | -0.1294 | No | ||
| 11 | SRPK2 | 5513 | 4466 | 0.095 | -0.1374 | No | ||
| 12 | GEMIN6 | 12492 7415 | 7235 | 0.013 | -0.2864 | No | ||
| 13 | LSM5 | 7286 | 7523 | 0.010 | -0.3018 | No | ||
| 14 | SLBP | 9822 | 10116 | -0.025 | -0.4411 | No | ||
| 15 | CRNKL1 | 14407 | 10520 | -0.031 | -0.4625 | No | ||
| 16 | CPSF3 | 7038 | 11079 | -0.040 | -0.4921 | No | ||
| 17 | GEMIN7 | 17944 | 13533 | -0.124 | -0.6228 | No | ||
| 18 | CUGBP1 | 2805 8819 4576 2924 | 14499 | -0.225 | -0.6721 | No | ||
| 19 | SFPQ | 12936 | 15076 | -0.341 | -0.6991 | No | ||
| 20 | SFRS1 | 8492 | 15244 | -0.391 | -0.7035 | No | ||
| 21 | KHDRBS1 | 5405 9778 | 15253 | -0.393 | -0.6993 | No | ||
| 22 | SNW1 | 7282 | 15575 | -0.516 | -0.7106 | No | ||
| 23 | KIN | 15120 | 15797 | -0.626 | -0.7151 | No | ||
| 24 | PRPF31 | 7594 | 16026 | -0.782 | -0.7182 | Yes | ||
| 25 | SFRS2 | 9807 20136 | 16049 | -0.798 | -0.7100 | Yes | ||
| 26 | NONO | 11993 | 16084 | -0.831 | -0.7021 | Yes | ||
| 27 | CSTF1 | 12515 | 16233 | -0.954 | -0.6988 | Yes | ||
| 28 | SFRS9 | 16731 | 16466 | -1.109 | -0.6983 | Yes | ||
| 29 | LSM3 | 12565 | 16490 | -1.130 | -0.6863 | Yes | ||
| 30 | SPOP | 5498 | 16515 | -1.153 | -0.6740 | Yes | ||
| 31 | TXNL4A | 6567 11329 | 16582 | -1.228 | -0.6631 | Yes | ||
| 32 | DHX15 | 8842 | 16588 | -1.232 | -0.6489 | Yes | ||
| 33 | NUDT21 | 12665 | 16589 | -1.233 | -0.6344 | Yes | ||
| 34 | RNMT | 7501 | 16629 | -1.276 | -0.6215 | Yes | ||
| 35 | PTBP1 | 5303 | 16652 | -1.298 | -0.6075 | Yes | ||
| 36 | SFRS5 | 9808 2062 | 16665 | -1.319 | -0.5926 | Yes | ||
| 37 | SF3A3 | 16091 | 17000 | -1.699 | -0.5906 | Yes | ||
| 38 | USP39 | 1116 1083 11373 | 17047 | -1.748 | -0.5726 | Yes | ||
| 39 | DHX38 | 48 | 17069 | -1.776 | -0.5528 | Yes | ||
| 40 | SF3A2 | 19938 | 17326 | -2.138 | -0.5415 | Yes | ||
| 41 | DDX39 | 18551 | 17362 | -2.193 | -0.5176 | Yes | ||
| 42 | SF3A1 | 7450 | 17384 | -2.247 | -0.4923 | Yes | ||
| 43 | DDX20 | 15213 | 17544 | -2.502 | -0.4715 | Yes | ||
| 44 | SFRS6 | 14751 | 17617 | -2.689 | -0.4437 | Yes | ||
| 45 | SIP1 | 21263 | 17619 | -2.696 | -0.4121 | Yes | ||
| 46 | FUSIP1 | 4715 16036 | 17627 | -2.708 | -0.3806 | Yes | ||
| 47 | PHF5A | 22194 | 17755 | -2.970 | -0.3526 | Yes | ||
| 48 | CPSF1 | 22241 | 17788 | -3.055 | -0.3184 | Yes | ||
| 49 | GEMIN5 | 20439 | 17808 | -3.106 | -0.2829 | Yes | ||
| 50 | CDC2L2 | 15964 2419 | 17856 | -3.204 | -0.2478 | Yes | ||
| 51 | SNRPD2 | 8412 | 17892 | -3.333 | -0.2105 | Yes | ||
| 52 | SNRPD1 | 23622 | 17897 | -3.342 | -0.1714 | Yes | ||
| 53 | SF3B3 | 18746 | 17955 | -3.510 | -0.1332 | Yes | ||
| 54 | EFTUD2 | 1219 20203 | 18188 | -4.255 | -0.0957 | Yes | ||
| 55 | SRPK1 | 23049 | 18311 | -4.799 | -0.0459 | Yes | ||
| 56 | GRSF1 | 16487 | 18393 | -5.299 | 0.0120 | Yes |