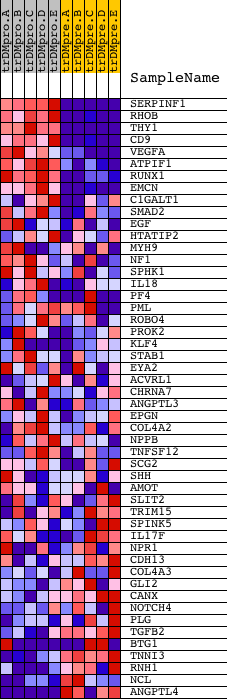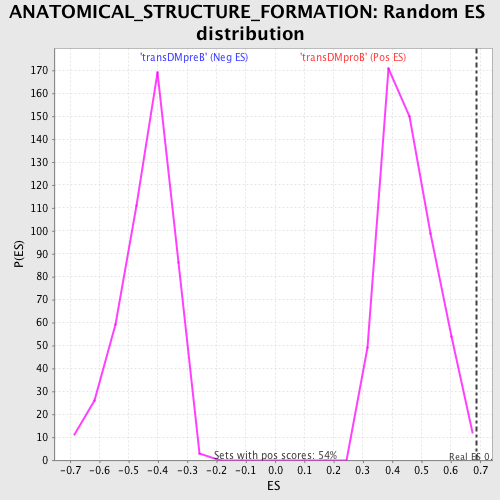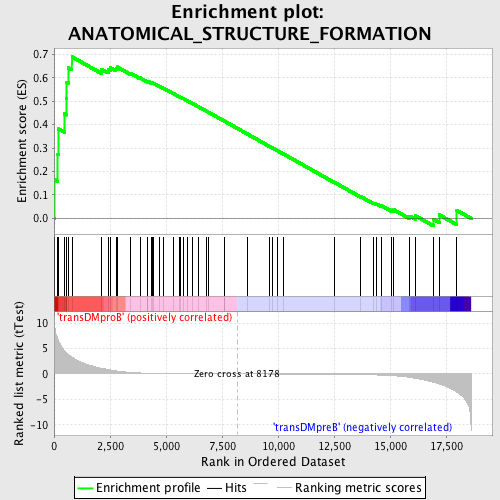
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | ANATOMICAL_STRUCTURE_FORMATION |
| Enrichment Score (ES) | 0.6891224 |
| Normalized Enrichment Score (NES) | 1.5186278 |
| Nominal p-value | 0.0037383179 |
| FDR q-value | 0.37677634 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SERPINF1 | 9793 | 18 | 9.741 | 0.1654 | Yes | ||
| 2 | RHOB | 4411 | 168 | 6.852 | 0.2743 | Yes | ||
| 3 | THY1 | 19481 | 199 | 6.383 | 0.3817 | Yes | ||
| 4 | CD9 | 16992 | 460 | 4.608 | 0.4464 | Yes | ||
| 5 | VEGFA | 22969 | 560 | 4.131 | 0.5116 | Yes | ||
| 6 | ATPIF1 | 4429 | 569 | 4.107 | 0.5813 | Yes | ||
| 7 | RUNX1 | 4481 | 640 | 3.781 | 0.6421 | Yes | ||
| 8 | EMCN | 7211 | 798 | 3.252 | 0.6891 | Yes | ||
| 9 | C1GALT1 | 8246 | 2101 | 1.071 | 0.6373 | No | ||
| 10 | SMAD2 | 23511 | 2417 | 0.844 | 0.6347 | No | ||
| 11 | EGF | 15169 | 2502 | 0.778 | 0.6435 | No | ||
| 12 | HTATIP2 | 18232 | 2762 | 0.581 | 0.6395 | No | ||
| 13 | MYH9 | 2252 2244 | 2813 | 0.548 | 0.6461 | No | ||
| 14 | NF1 | 5165 | 3387 | 0.293 | 0.6203 | No | ||
| 15 | SPHK1 | 5484 1266 1402 | 3834 | 0.183 | 0.5994 | No | ||
| 16 | IL18 | 9172 | 4165 | 0.128 | 0.5838 | No | ||
| 17 | PF4 | 16800 | 4170 | 0.128 | 0.5857 | No | ||
| 18 | PML | 3015 3074 3020 5270 | 4333 | 0.107 | 0.5789 | No | ||
| 19 | ROBO4 | 7895 3064 | 4370 | 0.104 | 0.5787 | No | ||
| 20 | PROK2 | 17057 | 4439 | 0.098 | 0.5767 | No | ||
| 21 | KLF4 | 9230 4962 4963 | 4692 | 0.079 | 0.5645 | No | ||
| 22 | STAB1 | 21892 | 4867 | 0.069 | 0.5563 | No | ||
| 23 | EYA2 | 2734 14729 | 5314 | 0.053 | 0.5332 | No | ||
| 24 | ACVRL1 | 22354 | 5579 | 0.045 | 0.5197 | No | ||
| 25 | CHRNA7 | 17808 | 5632 | 0.043 | 0.5176 | No | ||
| 26 | ANGPTL3 | 16171 | 5760 | 0.039 | 0.5115 | No | ||
| 27 | EPGN | 16798 | 5957 | 0.035 | 0.5015 | No | ||
| 28 | COL4A2 | 18688 | 6168 | 0.031 | 0.4907 | No | ||
| 29 | NPPB | 15994 | 6444 | 0.026 | 0.4764 | No | ||
| 30 | TNFSF12 | 10209 | 6801 | 0.020 | 0.4575 | No | ||
| 31 | SCG2 | 5410 14425 | 6887 | 0.018 | 0.4533 | No | ||
| 32 | SHH | 16581 | 7595 | 0.008 | 0.4153 | No | ||
| 33 | AMOT | 11341 24037 6587 | 7617 | 0.008 | 0.4143 | No | ||
| 34 | SLIT2 | 5456 | 8619 | -0.006 | 0.3605 | No | ||
| 35 | TRIM15 | 22988 | 9595 | -0.018 | 0.3083 | No | ||
| 36 | SPINK5 | 23569 | 9763 | -0.020 | 0.2997 | No | ||
| 37 | IL17F | 13992 | 9961 | -0.023 | 0.2894 | No | ||
| 38 | NPR1 | 9480 | 10234 | -0.026 | 0.2752 | No | ||
| 39 | CDH13 | 4506 3826 | 12518 | -0.077 | 0.1536 | No | ||
| 40 | COL4A3 | 8766 | 13684 | -0.136 | 0.0932 | No | ||
| 41 | GLI2 | 13859 | 14258 | -0.192 | 0.0656 | No | ||
| 42 | CANX | 4474 | 14411 | -0.212 | 0.0610 | No | ||
| 43 | NOTCH4 | 23277 9475 | 14621 | -0.244 | 0.0539 | No | ||
| 44 | PLG | 1593 23383 | 15062 | -0.337 | 0.0360 | No | ||
| 45 | TGFB2 | 5743 | 15170 | -0.369 | 0.0365 | No | ||
| 46 | BTG1 | 4458 4457 8663 | 15877 | -0.677 | 0.0100 | No | ||
| 47 | TNNI3 | 89 | 16129 | -0.869 | 0.0114 | No | ||
| 48 | RNH1 | 17562 | 16938 | -1.636 | -0.0042 | No | ||
| 49 | NCL | 5153 13899 | 17193 | -1.954 | 0.0155 | No | ||
| 50 | ANGPTL4 | 1504 7178 | 17979 | -3.580 | 0.0343 | No |