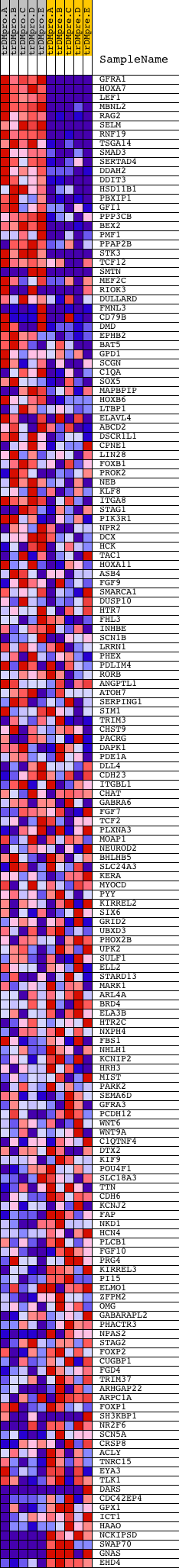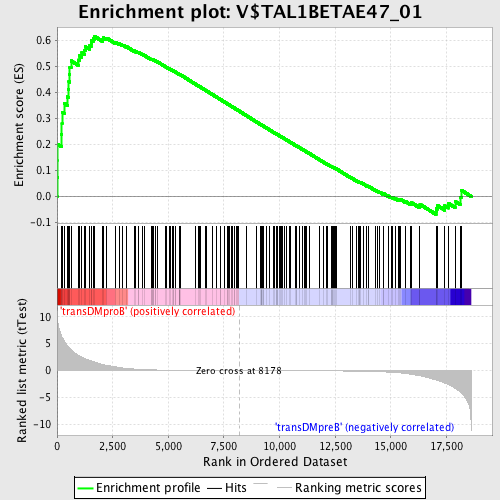
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$TAL1BETAE47_01 |
| Enrichment Score (ES) | 0.6176822 |
| Normalized Enrichment Score (NES) | 1.591589 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.051508002 |
| FWER p-Value | 0.318 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GFRA1 | 9015 | 7 | 10.510 | 0.0745 | Yes | ||
| 2 | HOXA7 | 4862 | 33 | 9.031 | 0.1375 | Yes | ||
| 3 | LEF1 | 1860 15420 | 37 | 8.921 | 0.2009 | Yes | ||
| 4 | MBNL2 | 21932 | 197 | 6.415 | 0.2380 | Yes | ||
| 5 | RAG2 | 14941 | 218 | 6.199 | 0.2811 | Yes | ||
| 6 | SELM | 20967 | 224 | 6.178 | 0.3249 | Yes | ||
| 7 | RNF19 | 22314 | 320 | 5.470 | 0.3587 | Yes | ||
| 8 | TSGA14 | 17194 | 462 | 4.590 | 0.3838 | Yes | ||
| 9 | SMAD3 | 19084 | 511 | 4.367 | 0.4123 | Yes | ||
| 10 | SERTAD4 | 13715 | 529 | 4.268 | 0.4418 | Yes | ||
| 11 | DDAH2 | 23263 | 553 | 4.149 | 0.4701 | Yes | ||
| 12 | DDIT3 | 19857 | 573 | 4.086 | 0.4982 | Yes | ||
| 13 | HSD11B1 | 9125 4019 | 631 | 3.850 | 0.5226 | Yes | ||
| 14 | PBXIP1 | 6028 | 950 | 2.867 | 0.5258 | Yes | ||
| 15 | GFI1 | 16451 | 1010 | 2.707 | 0.5419 | Yes | ||
| 16 | PPP3CB | 5285 | 1113 | 2.475 | 0.5540 | Yes | ||
| 17 | BEX2 | 24055 | 1231 | 2.244 | 0.5636 | Yes | ||
| 18 | PMF1 | 12452 | 1266 | 2.180 | 0.5773 | Yes | ||
| 19 | PPAP2B | 7503 16161 | 1458 | 1.875 | 0.5803 | Yes | ||
| 20 | STK3 | 7084 | 1530 | 1.776 | 0.5891 | Yes | ||
| 21 | TCF12 | 10044 3125 | 1547 | 1.753 | 0.6008 | Yes | ||
| 22 | SMTN | 20551 1446 | 1621 | 1.653 | 0.6086 | Yes | ||
| 23 | MEF2C | 3204 9378 | 1666 | 1.612 | 0.6177 | Yes | ||
| 24 | RIOK3 | 7354 12425 1973 | 2058 | 1.107 | 0.6044 | No | ||
| 25 | DULLARD | 20816 | 2067 | 1.102 | 0.6118 | No | ||
| 26 | FMNL3 | 22134 5866 | 2231 | 0.994 | 0.6101 | No | ||
| 27 | CD79B | 20185 1309 | 2617 | 0.685 | 0.5941 | No | ||
| 28 | DMD | 24295 2647 | 2796 | 0.556 | 0.5884 | No | ||
| 29 | EPHB2 | 4675 2440 8910 | 2951 | 0.476 | 0.5834 | No | ||
| 30 | BAT5 | 23261 | 3112 | 0.399 | 0.5776 | No | ||
| 31 | GPD1 | 22361 | 3456 | 0.271 | 0.5610 | No | ||
| 32 | SCGN | 3197 21516 | 3542 | 0.247 | 0.5581 | No | ||
| 33 | C1QA | 8666 | 3643 | 0.224 | 0.5543 | No | ||
| 34 | SOX5 | 9850 1044 5480 16937 | 3645 | 0.223 | 0.5558 | No | ||
| 35 | MAPBPIP | 15288 | 3835 | 0.183 | 0.5469 | No | ||
| 36 | HOXB6 | 20688 | 3910 | 0.168 | 0.5441 | No | ||
| 37 | LTBP1 | 1536 1604 6503 1559 1564 1518 | 4239 | 0.118 | 0.5271 | No | ||
| 38 | ELAVL4 | 15805 4889 9137 | 4250 | 0.116 | 0.5274 | No | ||
| 39 | ABCD2 | 11204 6494 | 4279 | 0.113 | 0.5267 | No | ||
| 40 | DSCR1L1 | 23231 | 4293 | 0.111 | 0.5268 | No | ||
| 41 | CPNE1 | 11186 | 4320 | 0.109 | 0.5262 | No | ||
| 42 | LIN28 | 15723 | 4329 | 0.108 | 0.5265 | No | ||
| 43 | FOXB1 | 12254 19069 | 4401 | 0.101 | 0.5234 | No | ||
| 44 | PROK2 | 17057 | 4439 | 0.098 | 0.5221 | No | ||
| 45 | NEB | 5158 | 4521 | 0.090 | 0.5183 | No | ||
| 46 | KLF8 | 10885 | 4866 | 0.069 | 0.5002 | No | ||
| 47 | ITGA8 | 14685 | 4926 | 0.066 | 0.4974 | No | ||
| 48 | STAG1 | 5520 9905 2987 | 5070 | 0.060 | 0.4901 | No | ||
| 49 | PIK3R1 | 3170 | 5108 | 0.059 | 0.4885 | No | ||
| 50 | NPR2 | 16230 | 5181 | 0.057 | 0.4850 | No | ||
| 51 | DCX | 24040 8841 4604 | 5211 | 0.056 | 0.4839 | No | ||
| 52 | HCK | 14787 | 5214 | 0.056 | 0.4842 | No | ||
| 53 | TAC1 | 17526 19853 | 5302 | 0.053 | 0.4798 | No | ||
| 54 | HOXA11 | 17146 | 5337 | 0.052 | 0.4784 | No | ||
| 55 | ASB4 | 990 17529 | 5490 | 0.047 | 0.4705 | No | ||
| 56 | FGF9 | 8966 | 5494 | 0.047 | 0.4706 | No | ||
| 57 | SMARCA1 | 24165 | 5497 | 0.047 | 0.4708 | No | ||
| 58 | DUSP10 | 4003 14016 | 5521 | 0.046 | 0.4699 | No | ||
| 59 | HTR7 | 23690 | 5535 | 0.046 | 0.4696 | No | ||
| 60 | FHL3 | 16092 2457 | 6240 | 0.029 | 0.4316 | No | ||
| 61 | INHBE | 19604 | 6365 | 0.027 | 0.4251 | No | ||
| 62 | SCN1B | 1148 17872 | 6389 | 0.026 | 0.4240 | No | ||
| 63 | LRRN1 | 17343 | 6438 | 0.026 | 0.4216 | No | ||
| 64 | PHEX | 24024 | 6679 | 0.022 | 0.4088 | No | ||
| 65 | PDLIM4 | 20455 | 6701 | 0.021 | 0.4078 | No | ||
| 66 | RORB | 10356 | 6976 | 0.017 | 0.3930 | No | ||
| 67 | ANGPTL1 | 10195 8589 13064 | 7163 | 0.014 | 0.3831 | No | ||
| 68 | ATOH7 | 20000 | 7348 | 0.012 | 0.3732 | No | ||
| 69 | SERPING1 | 14546 | 7356 | 0.012 | 0.3729 | No | ||
| 70 | SIM1 | 20031 | 7540 | 0.009 | 0.3630 | No | ||
| 71 | TRIM3 | 17703 | 7649 | 0.008 | 0.3572 | No | ||
| 72 | CHST9 | 7727 | 7668 | 0.007 | 0.3563 | No | ||
| 73 | PACRG | 23127 | 7714 | 0.007 | 0.3539 | No | ||
| 74 | DAPK1 | 7632 | 7753 | 0.006 | 0.3519 | No | ||
| 75 | PDE1A | 9541 5234 | 7817 | 0.005 | 0.3485 | No | ||
| 76 | DLL4 | 7040 | 7865 | 0.004 | 0.3460 | No | ||
| 77 | CDH23 | 3369 19759 3442 | 7976 | 0.003 | 0.3401 | No | ||
| 78 | ITGBL1 | 5850 | 7992 | 0.002 | 0.3393 | No | ||
| 79 | CHAT | 21882 | 8042 | 0.002 | 0.3366 | No | ||
| 80 | GABRA6 | 20491 | 8108 | 0.001 | 0.3331 | No | ||
| 81 | FGF7 | 8965 | 8131 | 0.001 | 0.3319 | No | ||
| 82 | TCF2 | 1395 20727 | 8517 | -0.004 | 0.3111 | No | ||
| 83 | PLXNA3 | 24299 | 8946 | -0.010 | 0.2880 | No | ||
| 84 | MOAP1 | 12252 | 9135 | -0.012 | 0.2779 | No | ||
| 85 | NEUROD2 | 20262 | 9169 | -0.012 | 0.2762 | No | ||
| 86 | BHLHB5 | 15629 | 9170 | -0.012 | 0.2763 | No | ||
| 87 | SLC24A3 | 14818 | 9175 | -0.013 | 0.2761 | No | ||
| 88 | KERA | 19895 | 9218 | -0.013 | 0.2739 | No | ||
| 89 | MYOCD | 1382 20406 1401 | 9282 | -0.014 | 0.2706 | No | ||
| 90 | PYY | 20207 | 9284 | -0.014 | 0.2707 | No | ||
| 91 | KIRREL2 | 17889 3922 | 9420 | -0.016 | 0.2635 | No | ||
| 92 | SIX6 | 21247 | 9558 | -0.017 | 0.2562 | No | ||
| 93 | GRID2 | 4803 | 9711 | -0.019 | 0.2481 | No | ||
| 94 | UBXD3 | 15701 | 9781 | -0.020 | 0.2445 | No | ||
| 95 | PHOX2B | 16524 | 9873 | -0.021 | 0.2397 | No | ||
| 96 | UPK2 | 19146 | 9882 | -0.022 | 0.2394 | No | ||
| 97 | SULF1 | 6285 | 9912 | -0.022 | 0.2380 | No | ||
| 98 | ELL2 | 5329 | 9920 | -0.022 | 0.2378 | No | ||
| 99 | STARD13 | 10833 | 9997 | -0.023 | 0.2338 | No | ||
| 100 | MARK1 | 5955 | 10040 | -0.024 | 0.2317 | No | ||
| 101 | ARL4A | 8627 | 10082 | -0.024 | 0.2297 | No | ||
| 102 | BRD4 | 7164 1609 1628 1598 1552 1601 12176 1630 1501 | 10124 | -0.025 | 0.2276 | No | ||
| 103 | ELA3B | 12592 7494 | 10221 | -0.026 | 0.2226 | No | ||
| 104 | HTR2C | 24230 | 10291 | -0.027 | 0.2191 | No | ||
| 105 | NXPH4 | 19602 | 10425 | -0.029 | 0.2121 | No | ||
| 106 | FBS1 | 18067 | 10510 | -0.031 | 0.2077 | No | ||
| 107 | NHLH1 | 13754 | 10724 | -0.034 | 0.1964 | No | ||
| 108 | KCNIP2 | 23655 3725 | 10757 | -0.035 | 0.1949 | No | ||
| 109 | HRH3 | 14317 | 10758 | -0.035 | 0.1952 | No | ||
| 110 | MIST | 16543 | 10913 | -0.037 | 0.1871 | No | ||
| 111 | PARK2 | 6944 11939 1499 23385 | 11051 | -0.040 | 0.1800 | No | ||
| 112 | SEMA6D | 2800 14876 90 | 11129 | -0.041 | 0.1761 | No | ||
| 113 | GFRA3 | 23469 | 11171 | -0.042 | 0.1742 | No | ||
| 114 | PCDH12 | 7005 | 11229 | -0.043 | 0.1714 | No | ||
| 115 | WNT6 | 5881 | 11353 | -0.046 | 0.1651 | No | ||
| 116 | WNT9A | 5709 | 11776 | -0.055 | 0.1426 | No | ||
| 117 | C1QTNF4 | 14953 | 11975 | -0.060 | 0.1323 | No | ||
| 118 | DTX2 | 16674 3488 | 12086 | -0.063 | 0.1268 | No | ||
| 119 | KIF9 | 19292 | 12172 | -0.066 | 0.1226 | No | ||
| 120 | POU4F1 | 21727 | 12317 | -0.070 | 0.1153 | No | ||
| 121 | SLC18A3 | 21883 | 12327 | -0.070 | 0.1153 | No | ||
| 122 | TTN | 14552 2743 14553 | 12382 | -0.072 | 0.1129 | No | ||
| 123 | CDH6 | 22320 | 12386 | -0.072 | 0.1133 | No | ||
| 124 | KCNJ2 | 20612 | 12406 | -0.073 | 0.1128 | No | ||
| 125 | FAP | 14577 | 12450 | -0.074 | 0.1110 | No | ||
| 126 | NKD1 | 8228 | 12492 | -0.076 | 0.1093 | No | ||
| 127 | HCN4 | 9080 | 12577 | -0.079 | 0.1053 | No | ||
| 128 | PLCB1 | 14832 2821 | 13167 | -0.103 | 0.0741 | No | ||
| 129 | FGF10 | 4719 8962 | 13285 | -0.109 | 0.0685 | No | ||
| 130 | PRG4 | 13592 4036 | 13455 | -0.120 | 0.0602 | No | ||
| 131 | KIRREL3 | 12571 | 13532 | -0.124 | 0.0570 | No | ||
| 132 | PI15 | 13586 | 13588 | -0.128 | 0.0549 | No | ||
| 133 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13606 | -0.130 | 0.0549 | No | ||
| 134 | ZFPM2 | 22483 | 13621 | -0.131 | 0.0551 | No | ||
| 135 | OMG | 5209 | 13750 | -0.141 | 0.0492 | No | ||
| 136 | GABARAPL2 | 8211 13552 | 13924 | -0.156 | 0.0409 | No | ||
| 137 | PHACTR3 | 7900 2770 | 13986 | -0.164 | 0.0388 | No | ||
| 138 | NPAS2 | 5186 | 14006 | -0.166 | 0.0390 | No | ||
| 139 | STAG2 | 5521 | 14304 | -0.198 | 0.0243 | No | ||
| 140 | FOXP2 | 4312 17523 8523 | 14407 | -0.211 | 0.0202 | No | ||
| 141 | CUGBP1 | 2805 8819 4576 2924 | 14499 | -0.225 | 0.0169 | No | ||
| 142 | FGD4 | 10306 5870 1717 1669 1700 | 14665 | -0.250 | 0.0098 | No | ||
| 143 | TRIM37 | 20715 | 14690 | -0.254 | 0.0103 | No | ||
| 144 | ARHGAP22 | 22050 | 14877 | -0.290 | 0.0023 | No | ||
| 145 | ARPC1A | 3541 16628 3519 | 15015 | -0.321 | -0.0029 | No | ||
| 146 | FOXP1 | 4242 | 15075 | -0.340 | -0.0036 | No | ||
| 147 | SH3KBP1 | 2631 12206 7189 | 15188 | -0.374 | -0.0071 | No | ||
| 148 | NR2F6 | 4651 8876 18848 8912 | 15344 | -0.425 | -0.0124 | No | ||
| 149 | SCN5A | 5412 | 15375 | -0.432 | -0.0110 | No | ||
| 150 | CRSP8 | 15062 | 15428 | -0.451 | -0.0106 | No | ||
| 151 | ACLY | 8348 | 15678 | -0.563 | -0.0201 | No | ||
| 152 | TNRC15 | 5971 | 15903 | -0.690 | -0.0273 | No | ||
| 153 | EYA3 | 8936 2352 | 15907 | -0.693 | -0.0225 | No | ||
| 154 | TLK1 | 14563 | 16293 | -0.995 | -0.0363 | No | ||
| 155 | DARS | 10375 13846 | 16309 | -0.999 | -0.0300 | No | ||
| 156 | CDC42EP4 | 20163 | 17034 | -1.731 | -0.0569 | No | ||
| 157 | GPX1 | 19310 | 17056 | -1.757 | -0.0455 | No | ||
| 158 | ICT1 | 20598 | 17109 | -1.816 | -0.0354 | No | ||
| 159 | HAAO | 22885 1578 | 17427 | -2.312 | -0.0360 | No | ||
| 160 | NCKIPSD | 19306 | 17580 | -2.586 | -0.0259 | No | ||
| 161 | SWAP70 | 18126 5553 9948 | 17890 | -3.322 | -0.0189 | No | ||
| 162 | GNAS | 9025 2963 2752 | 18118 | -3.994 | -0.0028 | No | ||
| 163 | EHD4 | 8275 | 18164 | -4.167 | 0.0245 | No |