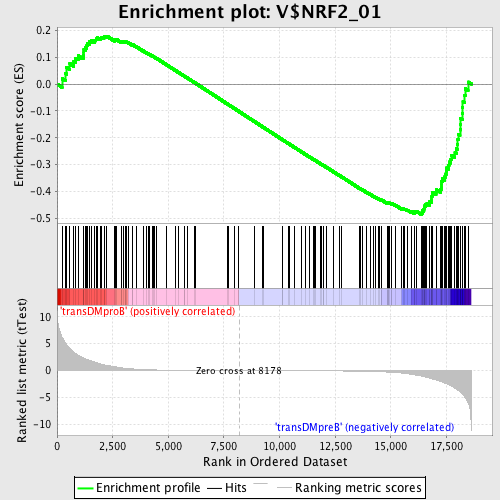
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$NRF2_01 |
| Enrichment Score (ES) | -0.4858497 |
| Normalized Enrichment Score (NES) | -1.2659608 |
| Nominal p-value | 0.038990825 |
| FDR q-value | 0.589008 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ITM2C | 14204 | 223 | 6.179 | 0.0206 | No | ||
| 2 | PRKACB | 15140 | 377 | 5.058 | 0.0391 | No | ||
| 3 | SCAMP2 | 19430 | 423 | 4.810 | 0.0622 | No | ||
| 4 | DDIT3 | 19857 | 573 | 4.086 | 0.0757 | No | ||
| 5 | RAP2C | 24156 | 751 | 3.382 | 0.0840 | No | ||
| 6 | TBCC | 23215 | 825 | 3.180 | 0.0969 | No | ||
| 7 | DPP8 | 19408 | 947 | 2.874 | 0.1056 | No | ||
| 8 | RRAS | 18256 | 1171 | 2.376 | 0.1061 | No | ||
| 9 | DAZAP2 | 10758 | 1199 | 2.316 | 0.1169 | No | ||
| 10 | ARHGAP4 | 9343 | 1204 | 2.301 | 0.1289 | No | ||
| 11 | FIBP | 7195 | 1264 | 2.185 | 0.1372 | No | ||
| 12 | CD2BP2 | 17621 | 1337 | 2.070 | 0.1443 | No | ||
| 13 | SEC24C | 22086 | 1374 | 1.997 | 0.1529 | No | ||
| 14 | FBXW9 | 18540 | 1460 | 1.874 | 0.1582 | No | ||
| 15 | UQCRH | 12378 | 1549 | 1.750 | 0.1627 | No | ||
| 16 | STK4 | 14743 | 1681 | 1.594 | 0.1641 | No | ||
| 17 | HMG20A | 19436 | 1747 | 1.498 | 0.1685 | No | ||
| 18 | NR1H2 | 17846 | 1792 | 1.433 | 0.1737 | No | ||
| 19 | PRDX5 | 7053 3732 | 1964 | 1.208 | 0.1708 | No | ||
| 20 | CPT2 | 15812 900 | 2006 | 1.162 | 0.1748 | No | ||
| 21 | CAP1 | 8684 | 2108 | 1.062 | 0.1749 | No | ||
| 22 | RGL1 | 9723 4072 | 2138 | 1.025 | 0.1788 | No | ||
| 23 | GTF3C1 | 10607 17646 | 2240 | 0.988 | 0.1785 | No | ||
| 24 | TBC1D17 | 6121 | 2580 | 0.715 | 0.1640 | No | ||
| 25 | AKAP12 | 17600 | 2616 | 0.686 | 0.1657 | No | ||
| 26 | EIF2B2 | 21204 | 2683 | 0.637 | 0.1655 | No | ||
| 27 | ARHGAP1 | 6001 10448 | 2873 | 0.518 | 0.1580 | No | ||
| 28 | ATP6V1D | 7872 | 2880 | 0.513 | 0.1604 | No | ||
| 29 | CSAD | 10899 | 2974 | 0.466 | 0.1578 | No | ||
| 30 | ARF3 | 22137 | 3053 | 0.430 | 0.1559 | No | ||
| 31 | RPS18 | 1529 5397 1603 | 3074 | 0.422 | 0.1570 | No | ||
| 32 | UBE1DC1 | 19020 | 3111 | 0.402 | 0.1572 | No | ||
| 33 | AP1GBP1 | 20725 | 3193 | 0.364 | 0.1548 | No | ||
| 34 | NF1 | 5165 | 3387 | 0.293 | 0.1459 | No | ||
| 35 | ELK4 | 8895 4664 3937 | 3395 | 0.291 | 0.1470 | No | ||
| 36 | VPS52 | 23287 1622 | 3570 | 0.240 | 0.1389 | No | ||
| 37 | STK11IP | 14212 | 3865 | 0.177 | 0.1239 | No | ||
| 38 | ZDHHC5 | 14547 | 4020 | 0.149 | 0.1163 | No | ||
| 39 | PSMD13 | 18019 3897 | 4092 | 0.138 | 0.1132 | No | ||
| 40 | ZNF22 | 12497 7417 | 4135 | 0.132 | 0.1116 | No | ||
| 41 | JARID1C | 5460 2563 | 4268 | 0.113 | 0.1051 | No | ||
| 42 | LIN28 | 15723 | 4329 | 0.108 | 0.1024 | No | ||
| 43 | MTF1 | 5128 9421 | 4380 | 0.103 | 0.1002 | No | ||
| 44 | CRB3 | 23176 | 4453 | 0.096 | 0.0969 | No | ||
| 45 | SHC1 | 9813 9812 5430 | 4928 | 0.066 | 0.0715 | No | ||
| 46 | RPS10 | 12464 | 5317 | 0.053 | 0.0508 | No | ||
| 47 | BCL2L1 | 4440 2930 8652 | 5461 | 0.048 | 0.0433 | No | ||
| 48 | HNRPD | 8645 | 5720 | 0.041 | 0.0296 | No | ||
| 49 | DR1 | 16762 | 5744 | 0.040 | 0.0285 | No | ||
| 50 | SYT5 | 17986 | 5849 | 0.038 | 0.0231 | No | ||
| 51 | ZBTB11 | 11309 | 6189 | 0.030 | 0.0049 | No | ||
| 52 | ACP2 | 4318 | 6221 | 0.030 | 0.0034 | No | ||
| 53 | PAFAH1B2 | 5221 9525 | 7651 | 0.008 | -0.0740 | No | ||
| 54 | RNPS1 | 9730 23361 | 7695 | 0.007 | -0.0763 | No | ||
| 55 | FBXO3 | 2662 14934 | 7952 | 0.003 | -0.0901 | No | ||
| 56 | DNAJC1 | 8854 | 8132 | 0.001 | -0.0998 | No | ||
| 57 | FBXO22 | 19442 | 8139 | 0.000 | -0.1001 | No | ||
| 58 | ACTR2 | 20523 | 8867 | -0.009 | -0.1395 | No | ||
| 59 | ACIN1 | 3319 2807 21832 | 9221 | -0.013 | -0.1585 | No | ||
| 60 | CLMN | 20988 2119 2104 | 9265 | -0.014 | -0.1608 | No | ||
| 61 | INSIG2 | 3944 4124 4073 | 10112 | -0.025 | -0.2064 | No | ||
| 62 | GNAI3 | 15198 | 10128 | -0.025 | -0.2071 | No | ||
| 63 | BAT4 | 8172 8173 | 10379 | -0.029 | -0.2205 | No | ||
| 64 | FMR1 | 24321 | 10441 | -0.030 | -0.2237 | No | ||
| 65 | VAX1 | 23636 | 10663 | -0.033 | -0.2354 | No | ||
| 66 | EGR2 | 8886 | 10983 | -0.039 | -0.2525 | No | ||
| 67 | ARPC5 | 12580 7487 | 11164 | -0.042 | -0.2620 | No | ||
| 68 | SLC6A14 | 12168 | 11337 | -0.046 | -0.2711 | No | ||
| 69 | LRRC1 | 19059 3129 | 11529 | -0.049 | -0.2812 | No | ||
| 70 | FYCO1 | 18952 | 11576 | -0.050 | -0.2834 | No | ||
| 71 | RPS5 | 18391 | 11634 | -0.052 | -0.2862 | No | ||
| 72 | FGFR1OP2 | 12544 12543 7457 | 11837 | -0.057 | -0.2969 | No | ||
| 73 | DDOST | 16021 | 11865 | -0.057 | -0.2980 | No | ||
| 74 | DRP2 | 24254 | 11971 | -0.060 | -0.3034 | No | ||
| 75 | GTF2A2 | 10654 | 11993 | -0.061 | -0.3042 | No | ||
| 76 | DTX2 | 16674 3488 | 12086 | -0.063 | -0.3089 | No | ||
| 77 | BIN3 | 21969 | 12437 | -0.074 | -0.3274 | No | ||
| 78 | DDX5 | 4606 8843 4607 | 12671 | -0.082 | -0.3396 | No | ||
| 79 | ARHGEF7 | 3823 | 12796 | -0.086 | -0.3459 | No | ||
| 80 | ING4 | 6602 1155 1102 11371 | 13580 | -0.128 | -0.3876 | No | ||
| 81 | DAXX | 23290 | 13639 | -0.133 | -0.3900 | No | ||
| 82 | RPL27 | 9740 | 13731 | -0.140 | -0.3942 | No | ||
| 83 | PHF6 | 2648 24340 | 13901 | -0.155 | -0.4025 | No | ||
| 84 | CSNK2B | 23008 | 14078 | -0.171 | -0.4112 | No | ||
| 85 | EIF2S1 | 4658 | 14218 | -0.187 | -0.4177 | No | ||
| 86 | TRIM44 | 8164 13487 | 14320 | -0.199 | -0.4221 | No | ||
| 87 | DIABLO | 16375 | 14436 | -0.215 | -0.4272 | No | ||
| 88 | CUGBP1 | 2805 8819 4576 2924 | 14499 | -0.225 | -0.4294 | No | ||
| 89 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 14565 | -0.235 | -0.4317 | No | ||
| 90 | HTATIP | 3690 | 14601 | -0.241 | -0.4323 | No | ||
| 91 | TNFRSF1A | 1181 10206 | 14854 | -0.284 | -0.4444 | No | ||
| 92 | USF1 | 5832 10260 10261 | 14870 | -0.287 | -0.4437 | No | ||
| 93 | RAB25 | 15287 | 14891 | -0.294 | -0.4432 | No | ||
| 94 | ZNF23 | 6149 | 14894 | -0.294 | -0.4418 | No | ||
| 95 | MTMR4 | 20712 | 14944 | -0.304 | -0.4428 | No | ||
| 96 | TCF7 | 1467 20466 | 15028 | -0.326 | -0.4456 | No | ||
| 97 | RPL37 | 12502 7421 22521 | 15051 | -0.333 | -0.4450 | No | ||
| 98 | ALX3 | 15456 | 15208 | -0.379 | -0.4515 | No | ||
| 99 | EPN1 | 18401 | 15486 | -0.476 | -0.4640 | No | ||
| 100 | TOMM70A | 6608 | 15553 | -0.508 | -0.4648 | No | ||
| 101 | IQGAP1 | 6619 | 15622 | -0.539 | -0.4657 | No | ||
| 102 | PHKB | 18537 3873 | 15749 | -0.604 | -0.4693 | No | ||
| 103 | B3GAT3 | 23938 | 15912 | -0.698 | -0.4744 | No | ||
| 104 | DHX8 | 20649 | 16062 | -0.808 | -0.4782 | No | ||
| 105 | NKIRAS2 | 12979 | 16064 | -0.809 | -0.4739 | No | ||
| 106 | PRKCSH | 19535 | 16143 | -0.879 | -0.4735 | No | ||
| 107 | AP1M1 | 18571 | 16372 | -1.027 | -0.4804 | Yes | ||
| 108 | MARK3 | 9369 | 16410 | -1.047 | -0.4769 | Yes | ||
| 109 | CBX8 | 11401 | 16414 | -1.049 | -0.4715 | Yes | ||
| 110 | DNMT1 | 19217 | 16473 | -1.117 | -0.4687 | Yes | ||
| 111 | SF3B4 | 22269 | 16492 | -1.131 | -0.4637 | Yes | ||
| 112 | ECGF1 | 22160 | 16512 | -1.147 | -0.4587 | Yes | ||
| 113 | ZNHIT1 | 16334 | 16532 | -1.181 | -0.4534 | Yes | ||
| 114 | ERCC1 | 1235 1045 1646 | 16562 | -1.213 | -0.4486 | Yes | ||
| 115 | NRAS | 5191 | 16623 | -1.268 | -0.4451 | Yes | ||
| 116 | UBE2N | 8216 19898 | 16734 | -1.407 | -0.4436 | Yes | ||
| 117 | AKT1S1 | 12557 | 16751 | -1.438 | -0.4369 | Yes | ||
| 118 | CUTC | 3711 | 16822 | -1.511 | -0.4327 | Yes | ||
| 119 | MTMR2 | 8069 3003 | 16839 | -1.537 | -0.4254 | Yes | ||
| 120 | HSPC171 | 18494 | 16842 | -1.538 | -0.4174 | Yes | ||
| 121 | ATBF1 | 916 8633 | 16857 | -1.556 | -0.4099 | Yes | ||
| 122 | EIF3S7 | 22226 | 16874 | -1.574 | -0.4024 | Yes | ||
| 123 | MTPN | 17183 | 17032 | -1.729 | -0.4018 | Yes | ||
| 124 | SENP1 | 10301 | 17041 | -1.737 | -0.3930 | Yes | ||
| 125 | COX15 | 5931 | 17225 | -1.998 | -0.3923 | Yes | ||
| 126 | UBE1 | 24370 2551 | 17268 | -2.066 | -0.3837 | Yes | ||
| 127 | RPL29 | 9741 | 17280 | -2.087 | -0.3732 | Yes | ||
| 128 | DLST | 8131 | 17289 | -2.096 | -0.3625 | Yes | ||
| 129 | RGS19 | 14303 | 17312 | -2.119 | -0.3525 | Yes | ||
| 130 | RPL6 | 9747 | 17417 | -2.298 | -0.3460 | Yes | ||
| 131 | TOMM40 | 6997 | 17444 | -2.345 | -0.3350 | Yes | ||
| 132 | CHERP | 11367 | 17487 | -2.430 | -0.3244 | Yes | ||
| 133 | TOMM22 | 5863 | 17509 | -2.462 | -0.3125 | Yes | ||
| 134 | PSMD12 | 20621 | 17591 | -2.622 | -0.3030 | Yes | ||
| 135 | SFRS6 | 14751 | 17617 | -2.689 | -0.2901 | Yes | ||
| 136 | MRPL3 | 3116 19335 | 17696 | -2.862 | -0.2792 | Yes | ||
| 137 | SIRT3 | 17569 3713 | 17730 | -2.921 | -0.2655 | Yes | ||
| 138 | TIMM8B | 11391 | 17883 | -3.299 | -0.2563 | Yes | ||
| 139 | DNAJC7 | 20227 | 17948 | -3.493 | -0.2412 | Yes | ||
| 140 | LZIC | 15988 | 17983 | -3.603 | -0.2240 | Yes | ||
| 141 | CHUK | 23665 | 18013 | -3.687 | -0.2060 | Yes | ||
| 142 | DDX50 | 3324 19749 | 18039 | -3.756 | -0.1875 | Yes | ||
| 143 | SDHD | 19125 | 18110 | -3.948 | -0.1704 | Yes | ||
| 144 | PLOD3 | 16666 | 18123 | -4.030 | -0.1497 | Yes | ||
| 145 | AAAS | 22111 2274 | 18134 | -4.076 | -0.1287 | Yes | ||
| 146 | GSPT1 | 9046 | 18209 | -4.329 | -0.1098 | Yes | ||
| 147 | TUFM | 10608 | 18227 | -4.435 | -0.0872 | Yes | ||
| 148 | NUDT5 | 15121 | 18243 | -4.469 | -0.0644 | Yes | ||
| 149 | KCNAB2 | 15655 | 18324 | -4.903 | -0.0427 | Yes | ||
| 150 | HSPH1 | 16282 | 18344 | -5.039 | -0.0171 | Yes | ||
| 151 | EIF5A | 11345 20379 6590 | 18471 | -5.997 | 0.0079 | Yes |