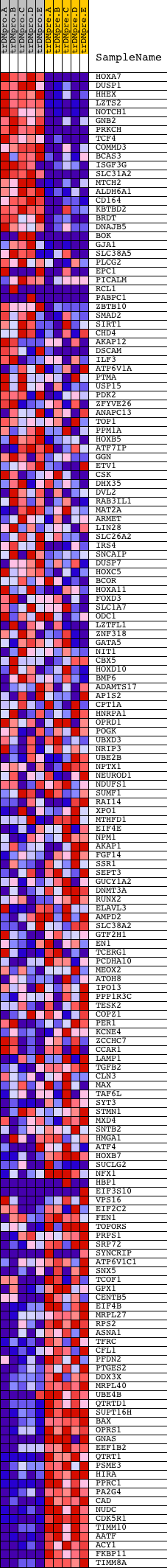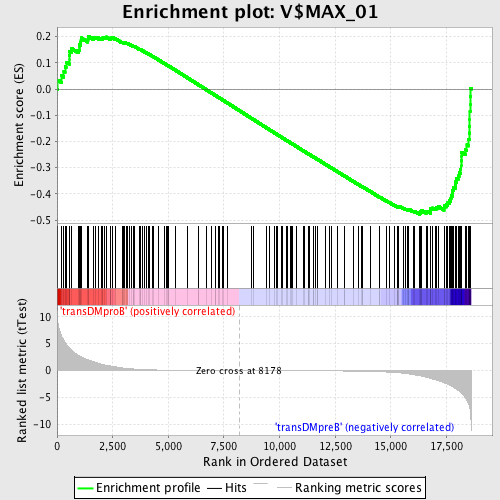
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$MAX_01 |
| Enrichment Score (ES) | -0.47860134 |
| Normalized Enrichment Score (NES) | -1.2794579 |
| Nominal p-value | 0.02173913 |
| FDR q-value | 0.60944426 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HOXA7 | 4862 | 33 | 9.031 | 0.0342 | No | ||
| 2 | DUSP1 | 23061 | 191 | 6.471 | 0.0514 | No | ||
| 3 | HHEX | 23872 | 306 | 5.589 | 0.0675 | No | ||
| 4 | LZTS2 | 10362 | 356 | 5.232 | 0.0857 | No | ||
| 5 | NOTCH1 | 14649 | 403 | 4.914 | 0.1028 | No | ||
| 6 | GNB2 | 9026 | 542 | 4.212 | 0.1121 | No | ||
| 7 | PRKCH | 21246 | 544 | 4.206 | 0.1287 | No | ||
| 8 | TCF4 | 5642 | 562 | 4.128 | 0.1443 | No | ||
| 9 | COMMD3 | 73 | 650 | 3.741 | 0.1544 | No | ||
| 10 | BCAS3 | 5314 | 961 | 2.839 | 0.1489 | No | ||
| 11 | ISGF3G | 2686 9184 | 992 | 2.774 | 0.1584 | No | ||
| 12 | SLC31A2 | 9827 | 995 | 2.759 | 0.1692 | No | ||
| 13 | MTCH2 | 14954 7120 | 1056 | 2.599 | 0.1763 | No | ||
| 14 | ALDH6A1 | 21025 | 1072 | 2.569 | 0.1858 | No | ||
| 15 | CD164 | 20046 | 1085 | 2.538 | 0.1952 | No | ||
| 16 | KBTBD2 | 17137 | 1379 | 1.991 | 0.1873 | No | ||
| 17 | BRDT | 4327 | 1389 | 1.972 | 0.1946 | No | ||
| 18 | DNAJB5 | 16236 | 1399 | 1.960 | 0.2019 | No | ||
| 19 | BOK | 11953 14174 | 1629 | 1.641 | 0.1961 | No | ||
| 20 | GJA1 | 20023 | 1705 | 1.562 | 0.1982 | No | ||
| 21 | SLC38A5 | 24388 | 1862 | 1.335 | 0.1951 | No | ||
| 22 | PLCG2 | 18453 | 1989 | 1.180 | 0.1929 | No | ||
| 23 | EPC1 | 2039 8907 2033 | 2057 | 1.107 | 0.1937 | No | ||
| 24 | PICALM | 18191 | 2059 | 1.107 | 0.1981 | No | ||
| 25 | RCL1 | 3728 3743 23894 | 2132 | 1.037 | 0.1983 | No | ||
| 26 | PABPC1 | 5219 9522 9523 23572 | 2200 | 1.000 | 0.1987 | No | ||
| 27 | ZBTB10 | 10468 | 2392 | 0.856 | 0.1917 | No | ||
| 28 | SMAD2 | 23511 | 2417 | 0.844 | 0.1938 | No | ||
| 29 | SIRT1 | 19745 | 2420 | 0.842 | 0.1970 | No | ||
| 30 | CHD4 | 8418 17281 4225 | 2472 | 0.796 | 0.1974 | No | ||
| 31 | AKAP12 | 17600 | 2616 | 0.686 | 0.1924 | No | ||
| 32 | DSCAM | 1667 4641 22530 | 2921 | 0.494 | 0.1779 | No | ||
| 33 | ILF3 | 3110 3030 9176 | 2949 | 0.476 | 0.1783 | No | ||
| 34 | ATP6V1A | 8638 | 3005 | 0.453 | 0.1771 | No | ||
| 35 | PTMA | 9657 | 3013 | 0.450 | 0.1786 | No | ||
| 36 | USP15 | 4758 3377 | 3102 | 0.407 | 0.1754 | No | ||
| 37 | PDK2 | 20285 | 3164 | 0.377 | 0.1736 | No | ||
| 38 | ZFYVE26 | 9999 | 3231 | 0.347 | 0.1714 | No | ||
| 39 | ANAPC13 | 12761 | 3346 | 0.305 | 0.1664 | No | ||
| 40 | TOP1 | 5790 5789 10210 | 3426 | 0.279 | 0.1633 | No | ||
| 41 | PPM1A | 5279 9607 | 3450 | 0.272 | 0.1631 | No | ||
| 42 | HOXB5 | 20687 | 3486 | 0.264 | 0.1623 | No | ||
| 43 | ATF7IP | 12028 13038 | 3684 | 0.215 | 0.1524 | No | ||
| 44 | GGN | 2472 1171 18312 | 3734 | 0.204 | 0.1506 | No | ||
| 45 | ETV1 | 4688 8925 8924 | 3759 | 0.198 | 0.1501 | No | ||
| 46 | CSK | 8805 | 3853 | 0.180 | 0.1458 | No | ||
| 47 | DHX35 | 14754 | 3944 | 0.162 | 0.1415 | No | ||
| 48 | DVL2 | 20813 | 4027 | 0.149 | 0.1377 | No | ||
| 49 | RAB3IL1 | 3723 13229 | 4107 | 0.136 | 0.1339 | No | ||
| 50 | MAT2A | 10553 10554 6099 | 4168 | 0.128 | 0.1312 | No | ||
| 51 | ARMET | 19005 | 4299 | 0.111 | 0.1246 | No | ||
| 52 | LIN28 | 15723 | 4329 | 0.108 | 0.1234 | No | ||
| 53 | SLC26A2 | 23427 | 4560 | 0.088 | 0.1113 | No | ||
| 54 | IRS4 | 9183 4926 | 4827 | 0.071 | 0.0972 | No | ||
| 55 | SNCAIP | 12589 | 4895 | 0.068 | 0.0938 | No | ||
| 56 | DUSP7 | 10661 | 4943 | 0.065 | 0.0915 | No | ||
| 57 | HOXC5 | 22340 | 4986 | 0.064 | 0.0895 | No | ||
| 58 | BCOR | 12934 2561 | 5027 | 0.062 | 0.0876 | No | ||
| 59 | HOXA11 | 17146 | 5337 | 0.052 | 0.0710 | No | ||
| 60 | FOXD3 | 9088 | 5838 | 0.038 | 0.0441 | No | ||
| 61 | SLC1A7 | 16147 | 6362 | 0.027 | 0.0158 | No | ||
| 62 | ODC1 | 9500 | 6373 | 0.027 | 0.0154 | No | ||
| 63 | LZTFL1 | 8210 | 6723 | 0.021 | -0.0034 | No | ||
| 64 | ZNF318 | 12198 7179 | 6950 | 0.017 | -0.0156 | No | ||
| 65 | GATA5 | 14315 | 7133 | 0.015 | -0.0254 | No | ||
| 66 | NIT1 | 11293 933 | 7134 | 0.015 | -0.0254 | No | ||
| 67 | CBX5 | 22108 4485 | 7268 | 0.013 | -0.0325 | No | ||
| 68 | HOXD10 | 14983 | 7287 | 0.013 | -0.0334 | No | ||
| 69 | BMP6 | 8659 | 7442 | 0.011 | -0.0417 | No | ||
| 70 | ADAMTS17 | 598 | 7492 | 0.010 | -0.0444 | No | ||
| 71 | AP1S2 | 8423 4228 | 7670 | 0.007 | -0.0539 | No | ||
| 72 | CPT1A | 23756 | 8719 | -0.007 | -0.1107 | No | ||
| 73 | HNRPA1 | 9103 4860 | 8807 | -0.008 | -0.1154 | No | ||
| 74 | OPRD1 | 15737 | 9423 | -0.016 | -0.1487 | No | ||
| 75 | POGK | 7739 | 9556 | -0.017 | -0.1558 | No | ||
| 76 | UBXD3 | 15701 | 9781 | -0.020 | -0.1678 | No | ||
| 77 | NRIP3 | 17679 | 9859 | -0.021 | -0.1719 | No | ||
| 78 | UBE2B | 10245 | 9919 | -0.022 | -0.1750 | No | ||
| 79 | NPTX1 | 20126 | 10100 | -0.025 | -0.1847 | No | ||
| 80 | NEUROD1 | 14550 | 10110 | -0.025 | -0.1851 | No | ||
| 81 | NDUFS1 | 13940 | 10315 | -0.028 | -0.1960 | No | ||
| 82 | SUMF1 | 17049 | 10333 | -0.028 | -0.1968 | No | ||
| 83 | RAI14 | 22327 2191 | 10502 | -0.031 | -0.2058 | No | ||
| 84 | XPO1 | 4172 | 10543 | -0.031 | -0.2079 | No | ||
| 85 | MTHFD1 | 2132 21238 | 10576 | -0.032 | -0.2095 | No | ||
| 86 | EIF4E | 15403 1827 8890 | 10781 | -0.035 | -0.2204 | No | ||
| 87 | NPM1 | 1196 | 11078 | -0.040 | -0.2363 | No | ||
| 88 | AKAP1 | 4364 1326 1286 20299 | 11108 | -0.041 | -0.2377 | No | ||
| 89 | FGF14 | 21716 3005 3463 | 11298 | -0.045 | -0.2477 | No | ||
| 90 | SSR1 | 4211 | 11316 | -0.045 | -0.2485 | No | ||
| 91 | SEPT3 | 6275 10775 | 11347 | -0.046 | -0.2499 | No | ||
| 92 | GUCY1A2 | 19580 | 11522 | -0.049 | -0.2592 | No | ||
| 93 | DNMT3A | 2167 21330 | 11537 | -0.050 | -0.2597 | No | ||
| 94 | RUNX2 | 4480 8700 | 11615 | -0.052 | -0.2637 | No | ||
| 95 | ELAVL3 | 9136 | 11714 | -0.053 | -0.2688 | No | ||
| 96 | AMPD2 | 1931 15197 | 11725 | -0.054 | -0.2691 | No | ||
| 97 | SLC38A2 | 22149 2219 | 12075 | -0.063 | -0.2878 | No | ||
| 98 | GTF2H1 | 4069 18236 | 12248 | -0.068 | -0.2968 | No | ||
| 99 | EN1 | 387 14155 | 12319 | -0.070 | -0.3004 | No | ||
| 100 | TCERG1 | 12075 2027 7068 | 12585 | -0.079 | -0.3144 | No | ||
| 101 | PCDHA10 | 8792 | 12902 | -0.091 | -0.3312 | No | ||
| 102 | MEOX2 | 21285 | 12913 | -0.092 | -0.3313 | No | ||
| 103 | ATOH8 | 17120 | 13342 | -0.111 | -0.3541 | No | ||
| 104 | IPO13 | 15784 | 13543 | -0.125 | -0.3644 | No | ||
| 105 | PPP1R3C | 23688 | 13697 | -0.137 | -0.3722 | No | ||
| 106 | TESK2 | 10504 | 13715 | -0.139 | -0.3726 | No | ||
| 107 | COPZ1 | 22338 | 13743 | -0.141 | -0.3735 | No | ||
| 108 | PER1 | 20827 | 14082 | -0.172 | -0.3911 | No | ||
| 109 | KCNE4 | 12196 | 14483 | -0.222 | -0.4119 | No | ||
| 110 | ZCCHC7 | 16225 | 14500 | -0.225 | -0.4119 | No | ||
| 111 | CCAR1 | 7454 | 14792 | -0.271 | -0.4266 | No | ||
| 112 | LAMP1 | 18679 | 14928 | -0.302 | -0.4327 | No | ||
| 113 | TGFB2 | 5743 | 15170 | -0.369 | -0.4443 | No | ||
| 114 | CLN3 | 17635 | 15299 | -0.410 | -0.4496 | No | ||
| 115 | MAX | 21034 | 15303 | -0.411 | -0.4481 | No | ||
| 116 | TAF6L | 5923 | 15321 | -0.418 | -0.4474 | No | ||
| 117 | SYT3 | 18264 | 15347 | -0.426 | -0.4470 | No | ||
| 118 | STMN1 | 9261 | 15365 | -0.429 | -0.4462 | No | ||
| 119 | MXD4 | 9346 | 15567 | -0.513 | -0.4551 | No | ||
| 120 | SNTB2 | 3887 18476 | 15680 | -0.564 | -0.4589 | No | ||
| 121 | HMGA1 | 23323 | 15756 | -0.608 | -0.4606 | No | ||
| 122 | ATF4 | 22417 2239 | 15807 | -0.634 | -0.4607 | No | ||
| 123 | HOXB7 | 666 9110 | 15812 | -0.637 | -0.4584 | No | ||
| 124 | SUCLG2 | 17062 5545 | 16000 | -0.766 | -0.4655 | No | ||
| 125 | NFX1 | 2475 | 16077 | -0.824 | -0.4664 | No | ||
| 126 | HBP1 | 2048 2162 21086 | 16304 | -0.998 | -0.4746 | Yes | ||
| 127 | EIF3S10 | 4659 8887 | 16329 | -1.005 | -0.4719 | Yes | ||
| 128 | VPS16 | 2927 14845 2825 | 16332 | -1.005 | -0.4680 | Yes | ||
| 129 | EIF2C2 | 6247 | 16389 | -1.034 | -0.4669 | Yes | ||
| 130 | FEN1 | 8961 | 16390 | -1.035 | -0.4628 | Yes | ||
| 131 | TOPORS | 15920 | 16586 | -1.232 | -0.4685 | Yes | ||
| 132 | PRPS1 | 24233 | 16633 | -1.283 | -0.4659 | Yes | ||
| 133 | SRP72 | 16819 | 16780 | -1.467 | -0.4680 | Yes | ||
| 134 | SYNCRIP | 3078 3035 3107 | 16783 | -1.471 | -0.4622 | Yes | ||
| 135 | ATP6V1C1 | 22487 12329 | 16790 | -1.477 | -0.4566 | Yes | ||
| 136 | SNX5 | 12783 14410 | 16852 | -1.553 | -0.4538 | Yes | ||
| 137 | TCOF1 | 5660 | 16995 | -1.697 | -0.4547 | Yes | ||
| 138 | GPX1 | 19310 | 17056 | -1.757 | -0.4510 | Yes | ||
| 139 | CENTB5 | 15961 | 17121 | -1.841 | -0.4471 | Yes | ||
| 140 | EIF4B | 13279 7979 | 17392 | -2.258 | -0.4527 | Yes | ||
| 141 | MRPL27 | 13573 | 17402 | -2.281 | -0.4442 | Yes | ||
| 142 | RPS2 | 9279 | 17512 | -2.464 | -0.4402 | Yes | ||
| 143 | ASNA1 | 18810 | 17565 | -2.554 | -0.4329 | Yes | ||
| 144 | TFRC | 22785 | 17647 | -2.739 | -0.4264 | Yes | ||
| 145 | CFL1 | 4516 | 17698 | -2.863 | -0.4177 | Yes | ||
| 146 | PFDN2 | 9551 | 17709 | -2.886 | -0.4067 | Yes | ||
| 147 | PTGES2 | 15038 | 17756 | -2.972 | -0.3974 | Yes | ||
| 148 | DDX3X | 24379 | 17785 | -3.050 | -0.3868 | Yes | ||
| 149 | MRPL40 | 22641 | 17822 | -3.132 | -0.3762 | Yes | ||
| 150 | UBE4B | 2433 15672 | 17896 | -3.342 | -0.3669 | Yes | ||
| 151 | QTRTD1 | 22590 | 17913 | -3.387 | -0.3543 | Yes | ||
| 152 | SUPT16H | 8541 | 17944 | -3.485 | -0.3420 | Yes | ||
| 153 | BAX | 17832 | 18024 | -3.718 | -0.3315 | Yes | ||
| 154 | OPRS1 | 9515 2402 | 18068 | -3.829 | -0.3186 | Yes | ||
| 155 | GNAS | 9025 2963 2752 | 18118 | -3.994 | -0.3053 | Yes | ||
| 156 | EEF1B2 | 4131 12063 | 18154 | -4.147 | -0.2907 | Yes | ||
| 157 | QTRT1 | 19540 | 18157 | -4.152 | -0.2743 | Yes | ||
| 158 | PSME3 | 20657 | 18163 | -4.167 | -0.2580 | Yes | ||
| 159 | HIRA | 4852 9090 | 18167 | -4.173 | -0.2415 | Yes | ||
| 160 | PPRC1 | 23808 | 18351 | -5.094 | -0.2312 | Yes | ||
| 161 | PA2G4 | 5265 | 18423 | -5.518 | -0.2130 | Yes | ||
| 162 | CAD | 16886 | 18503 | -6.237 | -0.1925 | Yes | ||
| 163 | NUDC | 9495 | 18520 | -6.456 | -0.1676 | Yes | ||
| 164 | CDK5R1 | 20745 | 18544 | -6.793 | -0.1418 | Yes | ||
| 165 | TIMM10 | 11390 2158 | 18553 | -6.965 | -0.1145 | Yes | ||
| 166 | AATF | 20315 | 18557 | -7.122 | -0.0863 | Yes | ||
| 167 | ACY1 | 19012 2996 | 18563 | -7.342 | -0.0574 | Yes | ||
| 168 | FKBP11 | 22136 | 18568 | -7.465 | -0.0279 | Yes | ||
| 169 | TIMM8A | 24062 | 18579 | -7.640 | 0.0020 | Yes |