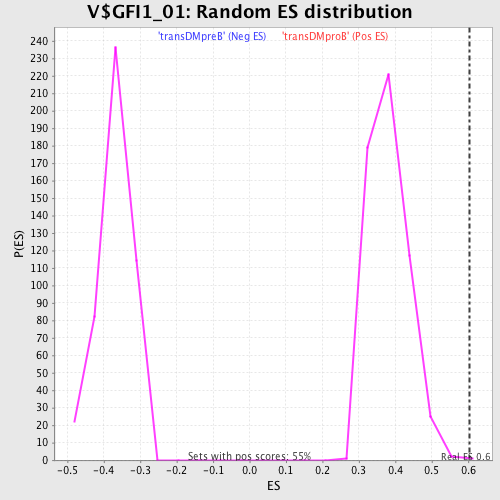
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$GFI1_01 |
| Enrichment Score (ES) | 0.60308903 |
| Normalized Enrichment Score (NES) | 1.5820819 |
| Nominal p-value | 0.0018315018 |
| FDR q-value | 0.048623588 |
| FWER p-Value | 0.369 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HOXA7 | 4862 | 33 | 9.031 | 0.0440 | Yes | ||
| 2 | PREP | 20035 | 101 | 7.625 | 0.0789 | Yes | ||
| 3 | IL16 | 9171 | 129 | 7.290 | 0.1144 | Yes | ||
| 4 | MEIS1 | 20524 | 179 | 6.639 | 0.1454 | Yes | ||
| 5 | GADD45A | 17129 | 196 | 6.435 | 0.1771 | Yes | ||
| 6 | SLC4A7 | 10181 | 214 | 6.243 | 0.2078 | Yes | ||
| 7 | SOX4 | 5479 | 232 | 6.101 | 0.2378 | Yes | ||
| 8 | MAP3K3 | 20626 | 266 | 5.885 | 0.2658 | Yes | ||
| 9 | BCL9L | 19475 | 267 | 5.879 | 0.2956 | Yes | ||
| 10 | RAB6B | 11289 | 321 | 5.467 | 0.3204 | Yes | ||
| 11 | TCTA | 4167 | 417 | 4.827 | 0.3397 | Yes | ||
| 12 | GABARAP | 20815 | 431 | 4.781 | 0.3632 | Yes | ||
| 13 | BCL6 | 22624 | 523 | 4.312 | 0.3801 | Yes | ||
| 14 | SERTAD4 | 13715 | 529 | 4.268 | 0.4015 | Yes | ||
| 15 | TCF4 | 5642 | 562 | 4.128 | 0.4207 | Yes | ||
| 16 | HSD11B1 | 9125 4019 | 631 | 3.850 | 0.4365 | Yes | ||
| 17 | PTEN | 5305 | 661 | 3.694 | 0.4536 | Yes | ||
| 18 | ABCF2 | 11338 6580 6579 | 743 | 3.425 | 0.4666 | Yes | ||
| 19 | MAP4K3 | 22887 | 754 | 3.376 | 0.4831 | Yes | ||
| 20 | SREBF2 | 5509 | 761 | 3.330 | 0.4997 | Yes | ||
| 21 | CAMK2D | 4232 | 775 | 3.295 | 0.5157 | Yes | ||
| 22 | PPARGC1A | 16533 | 835 | 3.159 | 0.5285 | Yes | ||
| 23 | TRAF4 | 10217 5796 1400 | 889 | 3.037 | 0.5410 | Yes | ||
| 24 | EXOC7 | 7003 | 912 | 2.980 | 0.5549 | Yes | ||
| 25 | PEA15 | 13753 | 982 | 2.790 | 0.5653 | Yes | ||
| 26 | IRF2BP1 | 18368 | 983 | 2.788 | 0.5794 | Yes | ||
| 27 | BMP4 | 21863 | 1004 | 2.727 | 0.5921 | Yes | ||
| 28 | MAP4K2 | 6457 | 1131 | 2.455 | 0.5977 | Yes | ||
| 29 | TCF12 | 10044 3125 | 1547 | 1.753 | 0.5841 | Yes | ||
| 30 | BCL9 | 15232 | 1579 | 1.709 | 0.5911 | Yes | ||
| 31 | BRD2 | 4735 8984 7628 | 1610 | 1.675 | 0.5979 | Yes | ||
| 32 | MEF2C | 3204 9378 | 1666 | 1.612 | 0.6031 | Yes | ||
| 33 | MAP2K5 | 19088 | 2016 | 1.152 | 0.5900 | No | ||
| 34 | CACNB2 | 4466 8678 | 2250 | 0.982 | 0.5823 | No | ||
| 35 | HNRPA0 | 13382 | 2262 | 0.973 | 0.5866 | No | ||
| 36 | CRYZL1 | 1644 12385 | 2473 | 0.794 | 0.5793 | No | ||
| 37 | WDR23 | 22009 | 2485 | 0.788 | 0.5827 | No | ||
| 38 | MYO1C | 20777 | 2655 | 0.661 | 0.5768 | No | ||
| 39 | HIVEP3 | 4973 | 2968 | 0.468 | 0.5623 | No | ||
| 40 | CSAD | 10899 | 2974 | 0.466 | 0.5644 | No | ||
| 41 | NFIA | 16172 5170 | 3020 | 0.446 | 0.5642 | No | ||
| 42 | HOXA9 | 17147 9109 1024 | 3024 | 0.446 | 0.5663 | No | ||
| 43 | TOP1 | 5790 5789 10210 | 3426 | 0.279 | 0.5459 | No | ||
| 44 | MBNL1 | 1921 15582 | 3427 | 0.279 | 0.5473 | No | ||
| 45 | LAMA3 | 23620 2009 | 3483 | 0.265 | 0.5457 | No | ||
| 46 | SOX5 | 9850 1044 5480 16937 | 3645 | 0.223 | 0.5381 | No | ||
| 47 | CTLA4 | 14238 | 3650 | 0.222 | 0.5390 | No | ||
| 48 | MYH10 | 8077 | 3781 | 0.195 | 0.5329 | No | ||
| 49 | PBX1 | 5224 | 3783 | 0.195 | 0.5339 | No | ||
| 50 | HOXB6 | 20688 | 3910 | 0.168 | 0.5279 | No | ||
| 51 | ANGPT1 | 8588 2260 22303 | 4099 | 0.137 | 0.5184 | No | ||
| 52 | ARHGEF2 | 4987 | 4115 | 0.134 | 0.5182 | No | ||
| 53 | ELAVL4 | 15805 4889 9137 | 4250 | 0.116 | 0.5115 | No | ||
| 54 | ZIC4 | 5992 | 4269 | 0.113 | 0.5111 | No | ||
| 55 | DSCR1L1 | 23231 | 4293 | 0.111 | 0.5105 | No | ||
| 56 | HIST1H1E | 21520 | 4319 | 0.109 | 0.5097 | No | ||
| 57 | SOX15 | 9848 | 4327 | 0.108 | 0.5098 | No | ||
| 58 | ZIC3 | 10430 | 4357 | 0.105 | 0.5088 | No | ||
| 59 | RORA | 3019 5388 | 4366 | 0.104 | 0.5089 | No | ||
| 60 | HIST1H1T | 13653 | 4461 | 0.095 | 0.5043 | No | ||
| 61 | TCF1 | 16416 | 4495 | 0.093 | 0.5029 | No | ||
| 62 | NGEF | 13891 | 4871 | 0.069 | 0.4829 | No | ||
| 63 | JMJD1C | 11531 19996 | 4975 | 0.064 | 0.4777 | No | ||
| 64 | DDR2 | 13767 | 5118 | 0.059 | 0.4703 | No | ||
| 65 | DCX | 24040 8841 4604 | 5211 | 0.056 | 0.4655 | No | ||
| 66 | DSPP | 8867 | 5331 | 0.052 | 0.4594 | No | ||
| 67 | ASB4 | 990 17529 | 5490 | 0.047 | 0.4510 | No | ||
| 68 | ICAM5 | 19543 | 5538 | 0.046 | 0.4487 | No | ||
| 69 | CREB5 | 10551 | 5982 | 0.035 | 0.4248 | No | ||
| 70 | TFEC | 17213 | 6482 | 0.025 | 0.3979 | No | ||
| 71 | NKX2-8 | 9465 | 6589 | 0.023 | 0.3922 | No | ||
| 72 | HOXA6 | 17150 | 6634 | 0.022 | 0.3900 | No | ||
| 73 | NCAM1 | 5149 | 6688 | 0.022 | 0.3872 | No | ||
| 74 | GREM1 | 14485 | 6772 | 0.020 | 0.3828 | No | ||
| 75 | NEXN | 12737 1903 | 6804 | 0.020 | 0.3812 | No | ||
| 76 | PDE4D | 10722 6235 | 6845 | 0.019 | 0.3791 | No | ||
| 77 | GAS2 | 4753 | 6870 | 0.019 | 0.3779 | No | ||
| 78 | BAI3 | 5580 | 6890 | 0.018 | 0.3770 | No | ||
| 79 | HOXA3 | 9108 1075 | 6953 | 0.017 | 0.3737 | No | ||
| 80 | PHYHIP | 21968 | 6963 | 0.017 | 0.3733 | No | ||
| 81 | HOXB8 | 9111 | 7203 | 0.014 | 0.3604 | No | ||
| 82 | PROCR | 14772 | 7240 | 0.013 | 0.3585 | No | ||
| 83 | PHF8 | 6770 | 7253 | 0.013 | 0.3579 | No | ||
| 84 | HOXD10 | 14983 | 7287 | 0.013 | 0.3562 | No | ||
| 85 | GPR20 | 22270 | 7475 | 0.010 | 0.3461 | No | ||
| 86 | LUZP1 | 6533 11257 2503 | 7571 | 0.009 | 0.3410 | No | ||
| 87 | CHRM1 | 23943 | 7972 | 0.003 | 0.3193 | No | ||
| 88 | CNKSR1 | 15719 20331 | 7982 | 0.003 | 0.3188 | No | ||
| 89 | TRDN | 20062 | 8068 | 0.001 | 0.3142 | No | ||
| 90 | TMPRSS2 | 22527 | 8357 | -0.003 | 0.2986 | No | ||
| 91 | MRGPRF | 17995 | 8366 | -0.003 | 0.2982 | No | ||
| 92 | SGK2 | 14750 | 8494 | -0.004 | 0.2913 | No | ||
| 93 | PURA | 9670 | 8758 | -0.007 | 0.2771 | No | ||
| 94 | CACNG3 | 12029 | 8907 | -0.009 | 0.2691 | No | ||
| 95 | CDON | 12195 | 8919 | -0.009 | 0.2685 | No | ||
| 96 | MSR1 | 5417 | 8921 | -0.009 | 0.2685 | No | ||
| 97 | NRXN3 | 2153 5196 | 9054 | -0.011 | 0.2614 | No | ||
| 98 | IRX3 | 18793 | 9252 | -0.013 | 0.2508 | No | ||
| 99 | CLMN | 20988 2119 2104 | 9265 | -0.014 | 0.2502 | No | ||
| 100 | GRPR | 24015 | 9432 | -0.016 | 0.2413 | No | ||
| 101 | MAP2K7 | 6453 | 9569 | -0.017 | 0.2340 | No | ||
| 102 | SLC26A3 | 21298 | 9588 | -0.018 | 0.2331 | No | ||
| 103 | EBF2 | 21975 | 9604 | -0.018 | 0.2324 | No | ||
| 104 | EVA1 | 19472 | 9732 | -0.020 | 0.2256 | No | ||
| 105 | JPH4 | 21826 | 9888 | -0.022 | 0.2173 | No | ||
| 106 | ICAM4 | 19544 | 10318 | -0.028 | 0.1941 | No | ||
| 107 | HOXA10 | 17145 989 | 10322 | -0.028 | 0.1941 | No | ||
| 108 | DEPDC2 | 14295 | 10421 | -0.029 | 0.1889 | No | ||
| 109 | ZIC1 | 19040 | 10540 | -0.031 | 0.1827 | No | ||
| 110 | CPEB3 | 5539 | 10625 | -0.033 | 0.1783 | No | ||
| 111 | TRIM23 | 8165 3230 | 10751 | -0.034 | 0.1717 | No | ||
| 112 | BPGM | 17489 | 10825 | -0.036 | 0.1679 | No | ||
| 113 | BDNF | 14926 2797 | 10908 | -0.037 | 0.1637 | No | ||
| 114 | GFRA3 | 23469 | 11171 | -0.042 | 0.1496 | No | ||
| 115 | DPF3 | 7659 12853 | 11187 | -0.042 | 0.1491 | No | ||
| 116 | RKHD3 | 18199 | 11443 | -0.048 | 0.1355 | No | ||
| 117 | OTP | 21582 | 11483 | -0.048 | 0.1336 | No | ||
| 118 | ITSN1 | 9196 | 11918 | -0.059 | 0.1103 | No | ||
| 119 | CGN | 12896 12897 7701 | 11955 | -0.060 | 0.1087 | No | ||
| 120 | ITGA10 | 15493 | 12014 | -0.061 | 0.1058 | No | ||
| 121 | PPP2R2A | 3222 21774 | 12043 | -0.062 | 0.1046 | No | ||
| 122 | CDH6 | 22320 | 12386 | -0.072 | 0.0864 | No | ||
| 123 | GPRC5D | 16962 | 12482 | -0.075 | 0.0817 | No | ||
| 124 | CDH13 | 4506 3826 | 12518 | -0.077 | 0.0801 | No | ||
| 125 | PCDH8 | 21739 3017 | 12672 | -0.082 | 0.0723 | No | ||
| 126 | BNC2 | 15850 | 12687 | -0.083 | 0.0719 | No | ||
| 127 | DSG3 | 8866 | 12745 | -0.085 | 0.0693 | No | ||
| 128 | EDN1 | 21658 | 12885 | -0.090 | 0.0622 | No | ||
| 129 | TPBG | 5793 | 13056 | -0.098 | 0.0534 | No | ||
| 130 | ETS1 | 10715 6230 3135 | 13060 | -0.098 | 0.0538 | No | ||
| 131 | POU3F1 | 16093 | 13085 | -0.099 | 0.0530 | No | ||
| 132 | RPS6KB1 | 7815 1207 13040 | 13099 | -0.100 | 0.0528 | No | ||
| 133 | KLF12 | 4960 2637 21732 9228 | 13177 | -0.104 | 0.0491 | No | ||
| 134 | RARB | 3011 21915 | 13214 | -0.106 | 0.0477 | No | ||
| 135 | HOXD11 | 9112 | 13237 | -0.107 | 0.0470 | No | ||
| 136 | CALN1 | 4713 | 13263 | -0.108 | 0.0462 | No | ||
| 137 | C1QTNF7 | 16852 | 13292 | -0.109 | 0.0453 | No | ||
| 138 | CNTN6 | 17346 | 13339 | -0.111 | 0.0433 | No | ||
| 139 | KLF15 | 17359 | 13505 | -0.123 | 0.0350 | No | ||
| 140 | PI15 | 13586 | 13588 | -0.128 | 0.0312 | No | ||
| 141 | ZFPM2 | 22483 | 13621 | -0.131 | 0.0301 | No | ||
| 142 | PTHLH | 16931 | 13801 | -0.145 | 0.0211 | No | ||
| 143 | PLCD4 | 9587 | 13804 | -0.146 | 0.0218 | No | ||
| 144 | PRIMA1 | 2055 20997 | 13819 | -0.148 | 0.0218 | No | ||
| 145 | PANK1 | 1687 23692 | 13858 | -0.151 | 0.0205 | No | ||
| 146 | ELMO3 | 10629 18495 | 13916 | -0.156 | 0.0182 | No | ||
| 147 | RHOA | 8624 4409 4410 | 13936 | -0.158 | 0.0179 | No | ||
| 148 | DNAJB4 | 12451 1916 1832 | 13966 | -0.161 | 0.0172 | No | ||
| 149 | PHACTR3 | 7900 2770 | 13986 | -0.164 | 0.0170 | No | ||
| 150 | TLR1 | 10190 | 14015 | -0.168 | 0.0163 | No | ||
| 151 | NR2F2 | 8619 409 | 14034 | -0.169 | 0.0162 | No | ||
| 152 | TSGA10 | 4123 13975 5587 | 14068 | -0.171 | 0.0153 | No | ||
| 153 | AVP | 14428 | 14134 | -0.177 | 0.0126 | No | ||
| 154 | SOX9 | 20611 | 14230 | -0.189 | 0.0084 | No | ||
| 155 | HOXB3 | 20685 | 14272 | -0.194 | 0.0072 | No | ||
| 156 | PDC | 4046 14102 | 14355 | -0.204 | 0.0038 | No | ||
| 157 | FOXP2 | 4312 17523 8523 | 14407 | -0.211 | 0.0021 | No | ||
| 158 | SP8 | 21122 | 14486 | -0.223 | -0.0010 | No | ||
| 159 | CDX2 | 16289 | 14568 | -0.236 | -0.0042 | No | ||
| 160 | GPR17 | 399 | 14599 | -0.240 | -0.0046 | No | ||
| 161 | ATF2 | 4418 2759 | 14872 | -0.288 | -0.0179 | No | ||
| 162 | MTMR4 | 20712 | 14944 | -0.304 | -0.0203 | No | ||
| 163 | CREM | 2005 1965 23496 1951 | 14953 | -0.306 | -0.0191 | No | ||
| 164 | SLC25A18 | 17303 | 15167 | -0.367 | -0.0288 | No | ||
| 165 | AGPAT4 | 23384 | 15209 | -0.379 | -0.0292 | No | ||
| 166 | HOXC8 | 4865 | 15332 | -0.421 | -0.0336 | No | ||
| 167 | STMN1 | 9261 | 15365 | -0.429 | -0.0332 | No | ||
| 168 | TBR1 | 83 | 15493 | -0.480 | -0.0377 | No | ||
| 169 | HOXC4 | 4864 | 15523 | -0.493 | -0.0367 | No | ||
| 170 | SOSTDC1 | 21286 | 15536 | -0.502 | -0.0349 | No | ||
| 171 | TFB1M | 23393 | 15617 | -0.533 | -0.0365 | No | ||
| 172 | HMGA1 | 23323 | 15756 | -0.608 | -0.0409 | No | ||
| 173 | SPAG7 | 20367 | 15831 | -0.646 | -0.0416 | No | ||
| 174 | POLH | 22966 | 15875 | -0.676 | -0.0406 | No | ||
| 175 | BTG1 | 4458 4457 8663 | 15877 | -0.677 | -0.0372 | No | ||
| 176 | ATP5G2 | 12610 | 16008 | -0.771 | -0.0403 | No | ||
| 177 | SFRS2 | 9807 20136 | 16049 | -0.798 | -0.0385 | No | ||
| 178 | TLK1 | 14563 | 16293 | -0.995 | -0.0466 | No | ||
| 179 | HBP1 | 2048 2162 21086 | 16304 | -0.998 | -0.0421 | No | ||
| 180 | BRUNOL4 | 2010 4229 | 16428 | -1.064 | -0.0434 | No | ||
| 181 | GRK6 | 6449 | 16456 | -1.100 | -0.0393 | No | ||
| 182 | MCTS1 | 24351 | 16593 | -1.236 | -0.0404 | No | ||
| 183 | YWHAE | 20776 | 16614 | -1.256 | -0.0351 | No | ||
| 184 | USP5 | 17004 | 16710 | -1.372 | -0.0333 | No | ||
| 185 | RKHD1 | 10693 | 16831 | -1.530 | -0.0321 | No | ||
| 186 | PRKAG1 | 22135 | 17363 | -2.193 | -0.0498 | No | ||
| 187 | ADD3 | 23812 3694 | 17439 | -2.337 | -0.0420 | No | ||
| 188 | SH3BGRL2 | 5601 | 17971 | -3.563 | -0.0528 | No | ||
| 189 | CCR7 | 20254 | 17975 | -3.570 | -0.0349 | No | ||
| 190 | PPARGC1B | 9318 | 17982 | -3.595 | -0.0170 | No | ||
| 191 | XPO5 | 7802 | 18082 | -3.855 | -0.0028 | No | ||
| 192 | PDLIM1 | 7023 | 18504 | -6.269 | 0.0061 | No |