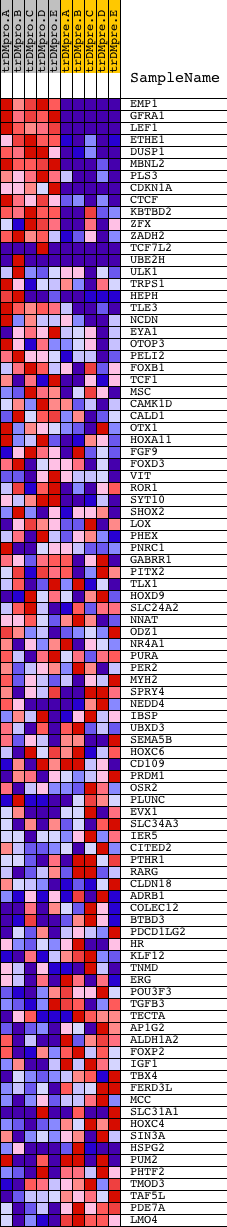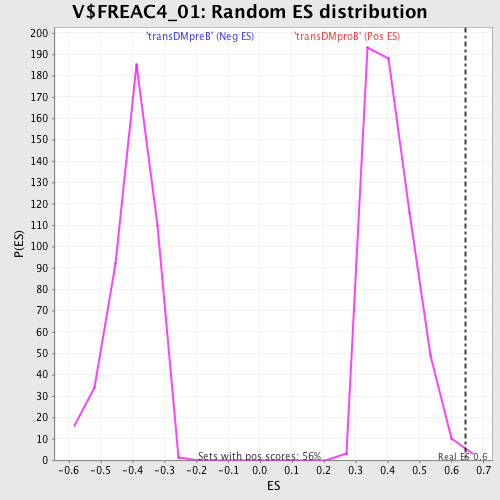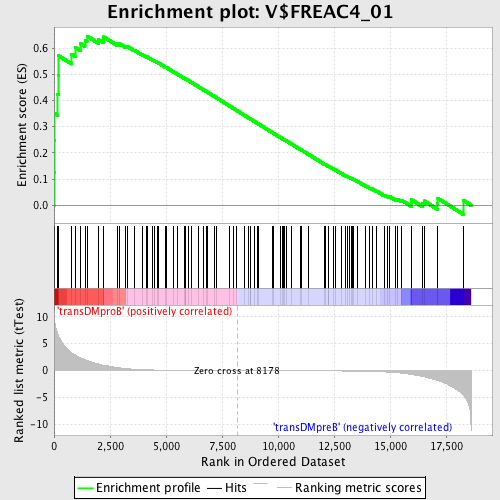
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$FREAC4_01 |
| Enrichment Score (ES) | 0.6458926 |
| Normalized Enrichment Score (NES) | 1.5735694 |
| Nominal p-value | 0.0035587188 |
| FDR q-value | 0.052137747 |
| FWER p-Value | 0.42 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 17260 | 2 | 10.883 | 0.1261 | Yes | ||
| 2 | GFRA1 | 9015 | 7 | 10.510 | 0.2477 | Yes | ||
| 3 | LEF1 | 1860 15420 | 37 | 8.921 | 0.3495 | Yes | ||
| 4 | ETHE1 | 18347 | 152 | 6.965 | 0.4241 | Yes | ||
| 5 | DUSP1 | 23061 | 191 | 6.471 | 0.4971 | Yes | ||
| 6 | MBNL2 | 21932 | 197 | 6.415 | 0.5711 | Yes | ||
| 7 | PLS3 | 8332 | 786 | 3.274 | 0.5774 | Yes | ||
| 8 | CDKN1A | 4511 8729 | 940 | 2.886 | 0.6026 | Yes | ||
| 9 | CTCF | 18490 | 1184 | 2.353 | 0.6167 | Yes | ||
| 10 | KBTBD2 | 17137 | 1379 | 1.991 | 0.6293 | Yes | ||
| 11 | ZFX | 5984 | 1471 | 1.853 | 0.6459 | Yes | ||
| 12 | ZADH2 | 23501 | 1988 | 1.181 | 0.6317 | No | ||
| 13 | TCF7L2 | 10048 5646 | 2198 | 1.001 | 0.6320 | No | ||
| 14 | UBE2H | 5823 5822 | 2215 | 0.998 | 0.6427 | No | ||
| 15 | ULK1 | 16436 | 2825 | 0.543 | 0.6162 | No | ||
| 16 | TRPS1 | 8195 | 2906 | 0.499 | 0.6176 | No | ||
| 17 | HEPH | 24288 2623 2635 2568 2652 | 3173 | 0.373 | 0.6076 | No | ||
| 18 | TLE3 | 5767 19416 | 3283 | 0.327 | 0.6055 | No | ||
| 19 | NCDN | 15756 2487 6471 | 3588 | 0.236 | 0.5918 | No | ||
| 20 | EYA1 | 4695 4061 | 3923 | 0.165 | 0.5757 | No | ||
| 21 | OTOP3 | 20599 | 4144 | 0.131 | 0.5654 | No | ||
| 22 | PELI2 | 8217 | 4146 | 0.131 | 0.5668 | No | ||
| 23 | FOXB1 | 12254 19069 | 4401 | 0.101 | 0.5543 | No | ||
| 24 | TCF1 | 16416 | 4495 | 0.093 | 0.5503 | No | ||
| 25 | MSC | 13998 | 4602 | 0.085 | 0.5456 | No | ||
| 26 | CAMK1D | 5977 2948 | 4665 | 0.081 | 0.5432 | No | ||
| 27 | CALD1 | 4273 8463 | 4983 | 0.064 | 0.5268 | No | ||
| 28 | OTX1 | 20517 | 4997 | 0.063 | 0.5268 | No | ||
| 29 | HOXA11 | 17146 | 5337 | 0.052 | 0.5091 | No | ||
| 30 | FGF9 | 8966 | 5494 | 0.047 | 0.5013 | No | ||
| 31 | FOXD3 | 9088 | 5838 | 0.038 | 0.4832 | No | ||
| 32 | VIT | 23155 | 5856 | 0.037 | 0.4827 | No | ||
| 33 | ROR1 | 6472 | 5860 | 0.037 | 0.4830 | No | ||
| 34 | SYT10 | 22157 | 5990 | 0.034 | 0.4764 | No | ||
| 35 | SHOX2 | 1802 5432 | 6154 | 0.031 | 0.4680 | No | ||
| 36 | LOX | 23438 | 6454 | 0.025 | 0.4521 | No | ||
| 37 | PHEX | 24024 | 6679 | 0.022 | 0.4403 | No | ||
| 38 | PNRC1 | 15925 | 6820 | 0.019 | 0.4329 | No | ||
| 39 | GABRR1 | 16250 | 6841 | 0.019 | 0.4321 | No | ||
| 40 | PITX2 | 15424 1878 | 7144 | 0.014 | 0.4159 | No | ||
| 41 | TLX1 | 23839 | 7238 | 0.013 | 0.4111 | No | ||
| 42 | HOXD9 | 14982 | 7821 | 0.005 | 0.3797 | No | ||
| 43 | SLC24A2 | 8020 13326 8021 | 8019 | 0.002 | 0.3691 | No | ||
| 44 | NNAT | 2764 5180 | 8129 | 0.001 | 0.3632 | No | ||
| 45 | ODZ1 | 24168 | 8484 | -0.004 | 0.3441 | No | ||
| 46 | NR4A1 | 9099 | 8659 | -0.006 | 0.3348 | No | ||
| 47 | PURA | 9670 | 8758 | -0.007 | 0.3296 | No | ||
| 48 | PER2 | 3940 13881 | 8769 | -0.008 | 0.3292 | No | ||
| 49 | MYH2 | 20838 | 8935 | -0.010 | 0.3204 | No | ||
| 50 | SPRY4 | 23448 | 9074 | -0.011 | 0.3131 | No | ||
| 51 | NEDD4 | 19384 3101 | 9122 | -0.012 | 0.3107 | No | ||
| 52 | IBSP | 16775 | 9764 | -0.020 | 0.2763 | No | ||
| 53 | UBXD3 | 15701 | 9781 | -0.020 | 0.2756 | No | ||
| 54 | SEMA5B | 9801 22777 | 10086 | -0.024 | 0.2595 | No | ||
| 55 | HOXC6 | 22339 2302 | 10194 | -0.026 | 0.2540 | No | ||
| 56 | CD109 | 6174 10656 19369 | 10224 | -0.026 | 0.2528 | No | ||
| 57 | PRDM1 | 19775 3337 | 10273 | -0.027 | 0.2505 | No | ||
| 58 | OSR2 | 22492 | 10392 | -0.029 | 0.2445 | No | ||
| 59 | PLUNC | 9593 | 10581 | -0.032 | 0.2347 | No | ||
| 60 | EVX1 | 17442 | 10988 | -0.039 | 0.2132 | No | ||
| 61 | SLC34A3 | 14664 | 11056 | -0.040 | 0.2101 | No | ||
| 62 | IER5 | 13802 | 11351 | -0.046 | 0.1947 | No | ||
| 63 | CITED2 | 5118 14477 | 12078 | -0.063 | 0.1563 | No | ||
| 64 | PTHR1 | 5318 18985 | 12124 | -0.064 | 0.1546 | No | ||
| 65 | RARG | 22113 | 12231 | -0.068 | 0.1496 | No | ||
| 66 | CLDN18 | 19028 | 12254 | -0.068 | 0.1492 | No | ||
| 67 | ADRB1 | 4357 | 12474 | -0.075 | 0.1383 | No | ||
| 68 | COLEC12 | 23624 8946 | 12550 | -0.078 | 0.1351 | No | ||
| 69 | BTBD3 | 10457 2975 6007 6006 | 12842 | -0.088 | 0.1205 | No | ||
| 70 | PDCD1LG2 | 23892 | 12995 | -0.095 | 0.1133 | No | ||
| 71 | HR | 3622 9119 | 13078 | -0.099 | 0.1101 | No | ||
| 72 | KLF12 | 4960 2637 21732 9228 | 13177 | -0.104 | 0.1060 | No | ||
| 73 | TNMD | 24260 | 13280 | -0.109 | 0.1017 | No | ||
| 74 | ERG | 1686 8915 | 13304 | -0.110 | 0.1018 | No | ||
| 75 | POU3F3 | 14261 | 13360 | -0.113 | 0.1001 | No | ||
| 76 | TGFB3 | 10161 | 13539 | -0.125 | 0.0919 | No | ||
| 77 | TECTA | 19159 | 13908 | -0.155 | 0.0739 | No | ||
| 78 | AP1G2 | 2677 21827 | 14095 | -0.173 | 0.0658 | No | ||
| 79 | ALDH1A2 | 19386 | 14199 | -0.185 | 0.0624 | No | ||
| 80 | FOXP2 | 4312 17523 8523 | 14407 | -0.211 | 0.0537 | No | ||
| 81 | IGF1 | 3352 9156 3409 | 14748 | -0.264 | 0.0384 | No | ||
| 82 | TBX4 | 5633 | 14876 | -0.290 | 0.0349 | No | ||
| 83 | FERD3L | 21292 | 14955 | -0.306 | 0.0342 | No | ||
| 84 | MCC | 552 | 15236 | -0.389 | 0.0236 | No | ||
| 85 | SLC31A1 | 2447 16195 | 15322 | -0.419 | 0.0239 | No | ||
| 86 | HOXC4 | 4864 | 15523 | -0.493 | 0.0188 | No | ||
| 87 | SIN3A | 5442 | 15952 | -0.728 | 0.0041 | No | ||
| 88 | HSPG2 | 4884 16023 | 15955 | -0.730 | 0.0125 | No | ||
| 89 | PUM2 | 2071 8161 | 15960 | -0.735 | 0.0208 | No | ||
| 90 | PHTF2 | 16599 | 16432 | -1.066 | 0.0077 | No | ||
| 91 | TMOD3 | 6945 | 16521 | -1.164 | 0.0164 | No | ||
| 92 | TAF5L | 18712 | 17123 | -1.844 | 0.0054 | No | ||
| 93 | PDE7A | 9544 1788 | 17134 | -1.859 | 0.0264 | No | ||
| 94 | LMO4 | 15151 | 18276 | -4.622 | 0.0184 | No |