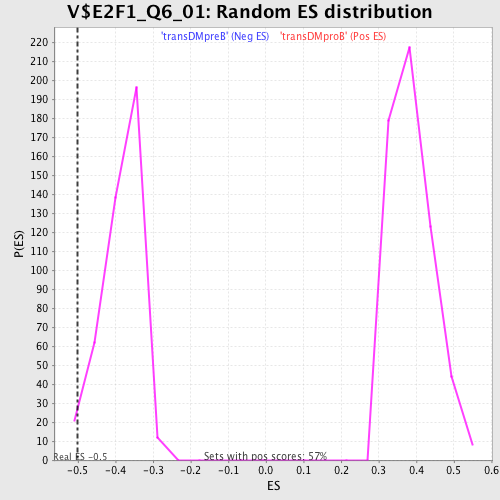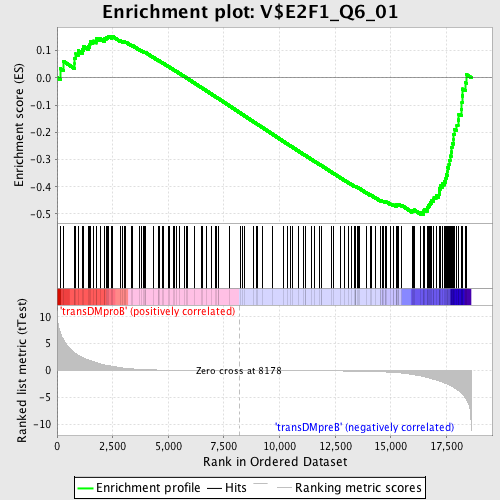
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$E2F1_Q6_01 |
| Enrichment Score (ES) | -0.50178885 |
| Normalized Enrichment Score (NES) | -1.3126488 |
| Nominal p-value | 0.02097902 |
| FDR q-value | 0.5355052 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GPRC5B | 17662 | 147 | 7.027 | 0.0337 | No | ||
| 2 | MXD3 | 21455 | 282 | 5.785 | 0.0607 | No | ||
| 3 | SLC25A1 | 22645 | 788 | 3.267 | 0.0527 | No | ||
| 4 | CASP2 | 8692 | 793 | 3.261 | 0.0718 | No | ||
| 5 | RAB11B | 9676 | 844 | 3.131 | 0.0877 | No | ||
| 6 | PPM1D | 20721 | 946 | 2.877 | 0.0992 | No | ||
| 7 | MCM4 | 22655 1708 | 1130 | 2.455 | 0.1039 | No | ||
| 8 | CTCF | 18490 | 1184 | 2.353 | 0.1149 | No | ||
| 9 | RBL1 | 14373 | 1404 | 1.956 | 0.1147 | No | ||
| 10 | ATE1 | 388 17602 | 1473 | 1.850 | 0.1219 | No | ||
| 11 | HIST1H2BK | 11423 | 1480 | 1.832 | 0.1325 | No | ||
| 12 | PIK3CD | 9563 | 1614 | 1.664 | 0.1351 | No | ||
| 13 | KLF5 | 4456 | 1751 | 1.491 | 0.1366 | No | ||
| 14 | FHOD1 | 18772 | 1761 | 1.477 | 0.1448 | No | ||
| 15 | HMGN2 | 9095 | 1929 | 1.257 | 0.1433 | No | ||
| 16 | NUMA1 | 18167 | 2136 | 1.027 | 0.1382 | No | ||
| 17 | TMPO | 5778 | 2142 | 1.024 | 0.1440 | No | ||
| 18 | RANBP1 | 9692 5357 | 2202 | 1.000 | 0.1467 | No | ||
| 19 | HIST1H2AH | 11414 | 2263 | 0.973 | 0.1492 | No | ||
| 20 | PHF13 | 15659 | 2318 | 0.914 | 0.1517 | No | ||
| 21 | AK2 | 8563 2479 16073 | 2445 | 0.817 | 0.1497 | No | ||
| 22 | CHD4 | 8418 17281 4225 | 2472 | 0.796 | 0.1530 | No | ||
| 23 | TBX6 | 1993 18075 | 2851 | 0.526 | 0.1357 | No | ||
| 24 | ILF3 | 3110 3030 9176 | 2949 | 0.476 | 0.1332 | No | ||
| 25 | PTMA | 9657 | 3013 | 0.450 | 0.1325 | No | ||
| 26 | MYC | 22465 9435 | 3081 | 0.419 | 0.1313 | No | ||
| 27 | USP52 | 3397 19841 | 3345 | 0.305 | 0.1189 | No | ||
| 28 | USP2 | 19480 3052 3043 | 3404 | 0.287 | 0.1175 | No | ||
| 29 | POU2F1 | 5275 3989 4065 4010 | 3707 | 0.209 | 0.1023 | No | ||
| 30 | MYH10 | 8077 | 3781 | 0.195 | 0.0995 | No | ||
| 31 | PHF15 | 20467 8050 | 3878 | 0.174 | 0.0954 | No | ||
| 32 | SP1 | 9852 | 3926 | 0.165 | 0.0938 | No | ||
| 33 | CCNT1 | 22140 11607 | 3966 | 0.159 | 0.0926 | No | ||
| 34 | ERF | 4680 8914 | 3970 | 0.158 | 0.0934 | No | ||
| 35 | UXT | 5837 10267 | 4331 | 0.107 | 0.0745 | No | ||
| 36 | NCOA6 | 2876 14382 | 4570 | 0.087 | 0.0622 | No | ||
| 37 | TNF | 23004 | 4572 | 0.087 | 0.0626 | No | ||
| 38 | HOXC10 | 22342 | 4601 | 0.085 | 0.0616 | No | ||
| 39 | HNRPUL1 | 6118 17923 1608 10577 1345 | 4716 | 0.078 | 0.0559 | No | ||
| 40 | DHH | 4629 | 4760 | 0.075 | 0.0540 | No | ||
| 41 | SYNGR4 | 17825 | 4993 | 0.063 | 0.0418 | No | ||
| 42 | STAG1 | 5520 9905 2987 | 5070 | 0.060 | 0.0380 | No | ||
| 43 | ADAMTS2 | 5706 20899 1356 | 5219 | 0.056 | 0.0304 | No | ||
| 44 | ARHGAP6 | 24207 | 5261 | 0.055 | 0.0285 | No | ||
| 45 | MGAT2 | 21256 | 5344 | 0.052 | 0.0243 | No | ||
| 46 | SMARCA1 | 24165 | 5497 | 0.047 | 0.0164 | No | ||
| 47 | HNRPD | 8645 | 5720 | 0.041 | 0.0046 | No | ||
| 48 | PCDH10 | 5227 9534 1893 | 5797 | 0.039 | 0.0007 | No | ||
| 49 | GNG4 | 21706 | 5877 | 0.037 | -0.0034 | No | ||
| 50 | SPHK2 | 1217 1806 12155 | 6194 | 0.030 | -0.0203 | No | ||
| 51 | EZH2 | 1092 17163 | 6473 | 0.025 | -0.0352 | No | ||
| 52 | CASP8AP2 | 16253 | 6550 | 0.024 | -0.0392 | No | ||
| 53 | TRIM39 | 22994 | 6710 | 0.021 | -0.0477 | No | ||
| 54 | ATM | 2976 19115 | 6926 | 0.018 | -0.0592 | No | ||
| 55 | STK35 | 14851 | 7098 | 0.015 | -0.0684 | No | ||
| 56 | FZD1 | 16923 | 7172 | 0.014 | -0.0723 | No | ||
| 57 | DNAJC9 | 21912 | 7233 | 0.013 | -0.0754 | No | ||
| 58 | CBX5 | 22108 4485 | 7268 | 0.013 | -0.0772 | No | ||
| 59 | EFNA5 | 4655 8884 | 7745 | 0.006 | -0.1030 | No | ||
| 60 | PODN | 15811 | 8257 | -0.001 | -0.1306 | No | ||
| 61 | MTSS1 | 5585 22285 | 8332 | -0.002 | -0.1346 | No | ||
| 62 | ATAD2 | 2268 2256 | 8408 | -0.003 | -0.1387 | No | ||
| 63 | HNRPA1 | 9103 4860 | 8807 | -0.008 | -0.1602 | No | ||
| 64 | RB1CC1 | 4486 14299 | 8978 | -0.010 | -0.1694 | No | ||
| 65 | PAX6 | 5223 | 9000 | -0.010 | -0.1704 | No | ||
| 66 | IRX3 | 18793 | 9252 | -0.013 | -0.1839 | No | ||
| 67 | NPAT | 19452 6356 | 9677 | -0.019 | -0.2068 | No | ||
| 68 | TYRO3 | 5811 | 10173 | -0.026 | -0.2335 | No | ||
| 69 | BMP2 | 14833 | 10347 | -0.028 | -0.2427 | No | ||
| 70 | HCN3 | 4843 1763 | 10499 | -0.030 | -0.2507 | No | ||
| 71 | PCSK1 | 21600 | 10565 | -0.032 | -0.2540 | No | ||
| 72 | POLE2 | 21053 | 10840 | -0.036 | -0.2686 | No | ||
| 73 | ORC1L | 327 16144 | 11076 | -0.040 | -0.2811 | No | ||
| 74 | LRP1 | 9284 | 11154 | -0.042 | -0.2851 | No | ||
| 75 | TOPBP1 | 10659 | 11174 | -0.042 | -0.2858 | No | ||
| 76 | RAD51 | 2897 14903 | 11436 | -0.047 | -0.2997 | No | ||
| 77 | SYT6 | 13437 7042 12040 | 11590 | -0.051 | -0.3077 | No | ||
| 78 | WEE1 | 18127 | 11778 | -0.055 | -0.3175 | No | ||
| 79 | YBX2 | 20818 | 11880 | -0.058 | -0.3226 | No | ||
| 80 | FAF1 | 2508 16137 8951 | 11893 | -0.058 | -0.3229 | No | ||
| 81 | CDC45L | 22642 1752 | 12352 | -0.071 | -0.3473 | No | ||
| 82 | FBXO5 | 19830 | 12426 | -0.073 | -0.3508 | No | ||
| 83 | ACTN3 | 23967 8540 | 12742 | -0.085 | -0.3674 | No | ||
| 84 | MCM6 | 4000 13845 4119 | 12928 | -0.092 | -0.3769 | No | ||
| 85 | HR | 3622 9119 | 13078 | -0.099 | -0.3844 | No | ||
| 86 | IPO7 | 6130 | 13229 | -0.106 | -0.3919 | No | ||
| 87 | FLI1 | 4729 | 13370 | -0.113 | -0.3988 | No | ||
| 88 | RPL18 | 450 5390 | 13396 | -0.115 | -0.3995 | No | ||
| 89 | MCM8 | 14834 | 13397 | -0.115 | -0.3988 | No | ||
| 90 | ERBB2IP | 3232 12234 | 13415 | -0.117 | -0.3990 | No | ||
| 91 | KCNS2 | 4949 2232 | 13487 | -0.121 | -0.4021 | No | ||
| 92 | PAQR4 | 23105 | 13540 | -0.125 | -0.4042 | No | ||
| 93 | REPS2 | 24018 | 13573 | -0.128 | -0.4052 | No | ||
| 94 | FANCC | 4712 | 13928 | -0.157 | -0.4234 | No | ||
| 95 | FKBP5 | 23051 | 14067 | -0.171 | -0.4299 | No | ||
| 96 | CNOT3 | 6112 10565 6113 | 14111 | -0.175 | -0.4312 | No | ||
| 97 | STAG2 | 5521 | 14304 | -0.198 | -0.4404 | No | ||
| 98 | TNFRSF19L | 11498 6727 6726 | 14515 | -0.227 | -0.4505 | No | ||
| 99 | HMGA2 | 4857 9098 3341 3444 | 14557 | -0.234 | -0.4513 | No | ||
| 100 | UFD1L | 22825 | 14625 | -0.244 | -0.4535 | No | ||
| 101 | ENPP1 | 19804 | 14644 | -0.248 | -0.4530 | No | ||
| 102 | PCNA | 9535 | 14678 | -0.252 | -0.4533 | No | ||
| 103 | HNRPR | 13196 7909 2500 | 14743 | -0.263 | -0.4552 | No | ||
| 104 | MAZ | 1327 17623 | 14779 | -0.268 | -0.4555 | No | ||
| 105 | FANCG | 15904 | 14820 | -0.276 | -0.4560 | No | ||
| 106 | PRKDC | 9617 22847 | 14988 | -0.313 | -0.4632 | No | ||
| 107 | DDB2 | 2721 2878 14525 2661 | 15126 | -0.357 | -0.4685 | No | ||
| 108 | AP4M1 | 3499 16656 | 15134 | -0.359 | -0.4668 | No | ||
| 109 | SFRS1 | 8492 | 15244 | -0.391 | -0.4704 | No | ||
| 110 | AP1S1 | 3500 3453 16335 | 15247 | -0.392 | -0.4681 | No | ||
| 111 | ASXL2 | 7957 | 15248 | -0.392 | -0.4658 | No | ||
| 112 | POLD3 | 17742 | 15292 | -0.408 | -0.4657 | No | ||
| 113 | GATA1 | 24196 | 15335 | -0.422 | -0.4655 | No | ||
| 114 | STMN1 | 9261 | 15365 | -0.429 | -0.4645 | No | ||
| 115 | JPH1 | 13994 | 15485 | -0.476 | -0.4682 | No | ||
| 116 | SIN3A | 5442 | 15952 | -0.728 | -0.4891 | No | ||
| 117 | ATP5G2 | 12610 | 16008 | -0.771 | -0.4875 | No | ||
| 118 | SFRS2 | 9807 20136 | 16049 | -0.798 | -0.4850 | No | ||
| 119 | E2F3 | 21500 8874 | 16351 | -1.017 | -0.4952 | Yes | ||
| 120 | DNMT1 | 19217 | 16473 | -1.117 | -0.4952 | Yes | ||
| 121 | DNAJC11 | 2486 6068 | 16479 | -1.120 | -0.4888 | Yes | ||
| 122 | ACBD6 | 14089 | 16525 | -1.168 | -0.4843 | Yes | ||
| 123 | PRPS1 | 24233 | 16633 | -1.283 | -0.4825 | Yes | ||
| 124 | MSH2 | 23138 | 16638 | -1.288 | -0.4751 | Yes | ||
| 125 | USP37 | 13923 | 16689 | -1.349 | -0.4698 | Yes | ||
| 126 | BRMS1L | 21268 | 16724 | -1.393 | -0.4634 | Yes | ||
| 127 | SYNCRIP | 3078 3035 3107 | 16783 | -1.471 | -0.4578 | Yes | ||
| 128 | CLSPN | 16079 11256 6530 | 16840 | -1.538 | -0.4517 | Yes | ||
| 129 | AMD1 | 8583 | 16910 | -1.614 | -0.4459 | Yes | ||
| 130 | ACO2 | 8527 | 16939 | -1.637 | -0.4377 | Yes | ||
| 131 | HTF9C | 4886 1678 | 17033 | -1.730 | -0.4325 | Yes | ||
| 132 | MCM7 | 9372 3568 | 17165 | -1.899 | -0.4284 | Yes | ||
| 133 | INSM1 | 14815 | 17177 | -1.916 | -0.4176 | Yes | ||
| 134 | NCL | 5153 13899 | 17193 | -1.954 | -0.4068 | Yes | ||
| 135 | NASP | 2383 2387 6955 | 17226 | -1.999 | -0.3967 | Yes | ||
| 136 | CDCA7 | 12437 | 17313 | -2.120 | -0.3888 | Yes | ||
| 137 | SLC9A7 | 24177 | 17403 | -2.283 | -0.3801 | Yes | ||
| 138 | RPS19 | 5398 | 17473 | -2.393 | -0.3697 | Yes | ||
| 139 | CDC5L | 7746 7745 7744 | 17489 | -2.432 | -0.3561 | Yes | ||
| 140 | RQCD1 | 12205 | 17549 | -2.514 | -0.3444 | Yes | ||
| 141 | MCM2 | 17074 | 17557 | -2.531 | -0.3298 | Yes | ||
| 142 | EED | 17759 | 17611 | -2.674 | -0.3168 | Yes | ||
| 143 | SUV39H1 | 24194 | 17637 | -2.723 | -0.3020 | Yes | ||
| 144 | SUMO1 | 5826 3943 10247 | 17665 | -2.792 | -0.2869 | Yes | ||
| 145 | MCM3 | 13991 | 17710 | -2.887 | -0.2722 | Yes | ||
| 146 | GMNN | 21513 | 17724 | -2.911 | -0.2557 | Yes | ||
| 147 | PHF5A | 22194 | 17755 | -2.970 | -0.2397 | Yes | ||
| 148 | MRPL40 | 22641 | 17822 | -3.132 | -0.2247 | Yes | ||
| 149 | CDC25A | 8721 | 17829 | -3.139 | -0.2064 | Yes | ||
| 150 | GADD45B | 19935 | 17855 | -3.198 | -0.1888 | Yes | ||
| 151 | NOLC1 | 7704 | 17970 | -3.562 | -0.1739 | Yes | ||
| 152 | E2F1 | 14384 | 18022 | -3.710 | -0.1547 | Yes | ||
| 153 | PPP1R8 | 15730 | 18057 | -3.802 | -0.1340 | Yes | ||
| 154 | HIRA | 4852 9090 | 18167 | -4.173 | -0.1152 | Yes | ||
| 155 | POLA2 | 23988 | 18195 | -4.278 | -0.0913 | Yes | ||
| 156 | GSPT1 | 9046 | 18209 | -4.329 | -0.0663 | Yes | ||
| 157 | UNG | 10257 16744 | 18224 | -4.409 | -0.0410 | Yes | ||
| 158 | PPRC1 | 23808 | 18351 | -5.094 | -0.0176 | Yes | ||
| 159 | MAP4K1 | 18313 | 18407 | -5.384 | 0.0113 | Yes |