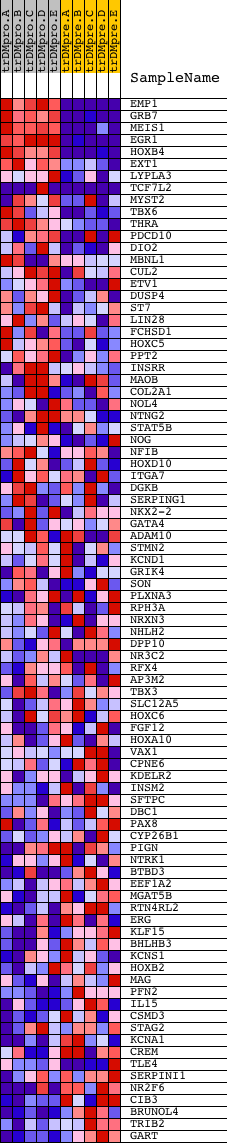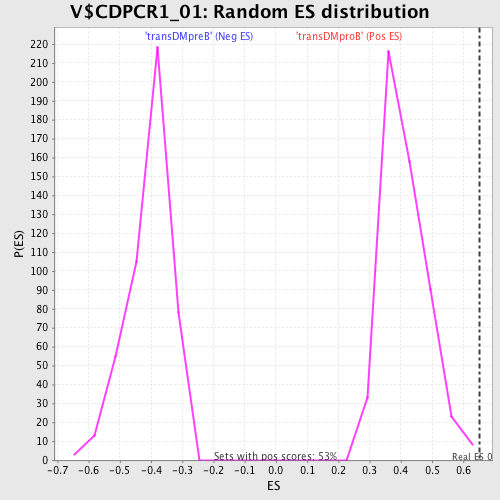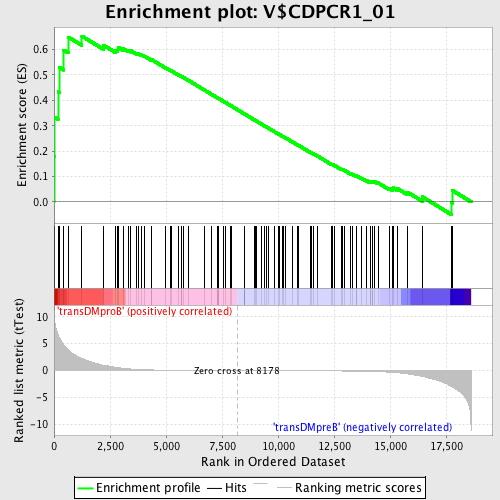
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$CDPCR1_01 |
| Enrichment Score (ES) | 0.65316224 |
| Normalized Enrichment Score (NES) | 1.5880079 |
| Nominal p-value | 0.0018939395 |
| FDR q-value | 0.047107376 |
| FWER p-Value | 0.337 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 17260 | 2 | 10.883 | 0.1781 | Yes | ||
| 2 | GRB7 | 20673 | 22 | 9.600 | 0.3342 | Yes | ||
| 3 | MEIS1 | 20524 | 179 | 6.639 | 0.4345 | Yes | ||
| 4 | EGR1 | 23598 | 249 | 5.993 | 0.5289 | Yes | ||
| 5 | HOXB4 | 20686 | 426 | 4.798 | 0.5980 | Yes | ||
| 6 | EXT1 | 22297 | 634 | 3.820 | 0.6494 | Yes | ||
| 7 | LYPLA3 | 18482 | 1240 | 2.226 | 0.6532 | Yes | ||
| 8 | TCF7L2 | 10048 5646 | 2198 | 1.001 | 0.6179 | No | ||
| 9 | MYST2 | 20283 | 2752 | 0.589 | 0.5977 | No | ||
| 10 | TBX6 | 1993 18075 | 2851 | 0.526 | 0.6010 | No | ||
| 11 | THRA | 1447 10171 1406 | 2857 | 0.523 | 0.6093 | No | ||
| 12 | PDCD10 | 15314 | 3101 | 0.407 | 0.6029 | No | ||
| 13 | DIO2 | 21014 2159 | 3324 | 0.312 | 0.5960 | No | ||
| 14 | MBNL1 | 1921 15582 | 3427 | 0.279 | 0.5951 | No | ||
| 15 | CUL2 | 23630 | 3679 | 0.215 | 0.5850 | No | ||
| 16 | ETV1 | 4688 8925 8924 | 3759 | 0.198 | 0.5840 | No | ||
| 17 | DUSP4 | 18632 3820 | 3906 | 0.168 | 0.5789 | No | ||
| 18 | ST7 | 1054 17519 | 4051 | 0.145 | 0.5735 | No | ||
| 19 | LIN28 | 15723 | 4329 | 0.108 | 0.5603 | No | ||
| 20 | FCHSD1 | 6661 11435 | 4336 | 0.107 | 0.5617 | No | ||
| 21 | HOXC5 | 22340 | 4986 | 0.064 | 0.5278 | No | ||
| 22 | PPT2 | 1617 12034 | 5209 | 0.056 | 0.5167 | No | ||
| 23 | INSRR | 15557 | 5255 | 0.055 | 0.5152 | No | ||
| 24 | MAOB | 24182 2556 | 5545 | 0.045 | 0.5003 | No | ||
| 25 | COL2A1 | 22145 4542 | 5665 | 0.042 | 0.4946 | No | ||
| 26 | NOL4 | 2028 11432 | 5682 | 0.042 | 0.4944 | No | ||
| 27 | NTNG2 | 2731 9330 | 5770 | 0.039 | 0.4903 | No | ||
| 28 | STAT5B | 20222 | 5983 | 0.035 | 0.4795 | No | ||
| 29 | NOG | 13663 | 6715 | 0.021 | 0.4404 | No | ||
| 30 | NFIB | 15855 | 7040 | 0.016 | 0.4231 | No | ||
| 31 | HOXD10 | 14983 | 7287 | 0.013 | 0.4101 | No | ||
| 32 | ITGA7 | 19835 | 7303 | 0.012 | 0.4095 | No | ||
| 33 | DGKB | 5722 | 7323 | 0.012 | 0.4086 | No | ||
| 34 | SERPING1 | 14546 | 7356 | 0.012 | 0.4071 | No | ||
| 35 | NKX2-2 | 5176 | 7542 | 0.009 | 0.3973 | No | ||
| 36 | GATA4 | 21792 4755 | 7650 | 0.008 | 0.3916 | No | ||
| 37 | ADAM10 | 4336 | 7875 | 0.004 | 0.3796 | No | ||
| 38 | STMN2 | 15638 | 7898 | 0.004 | 0.3785 | No | ||
| 39 | KCND1 | 4940 | 8477 | -0.004 | 0.3473 | No | ||
| 40 | GRIK4 | 19157 | 8498 | -0.004 | 0.3463 | No | ||
| 41 | SON | 5473 1657 1684 | 8509 | -0.004 | 0.3459 | No | ||
| 42 | PLXNA3 | 24299 | 8946 | -0.010 | 0.3225 | No | ||
| 43 | RPH3A | 13628 | 9009 | -0.010 | 0.3193 | No | ||
| 44 | NRXN3 | 2153 5196 | 9054 | -0.011 | 0.3171 | No | ||
| 45 | NHLH2 | 9462 | 9251 | -0.013 | 0.3068 | No | ||
| 46 | DPP10 | 13850 | 9376 | -0.015 | 0.3003 | No | ||
| 47 | NR3C2 | 18562 13798 | 9491 | -0.016 | 0.2944 | No | ||
| 48 | RFX4 | 7721 3410 | 9567 | -0.017 | 0.2907 | No | ||
| 49 | AP3M2 | 11 | 9824 | -0.021 | 0.2772 | No | ||
| 50 | TBX3 | 16723 | 10010 | -0.023 | 0.2676 | No | ||
| 51 | SLC12A5 | 14731 | 10062 | -0.024 | 0.2652 | No | ||
| 52 | HOXC6 | 22339 2302 | 10194 | -0.026 | 0.2586 | No | ||
| 53 | FGF12 | 1723 22621 | 10239 | -0.026 | 0.2566 | No | ||
| 54 | HOXA10 | 17145 989 | 10322 | -0.028 | 0.2527 | No | ||
| 55 | VAX1 | 23636 | 10663 | -0.033 | 0.2349 | No | ||
| 56 | CPNE6 | 22010 | 10883 | -0.037 | 0.2236 | No | ||
| 57 | KDELR2 | 16634 12428 | 10890 | -0.037 | 0.2239 | No | ||
| 58 | INSM2 | 19821 | 11449 | -0.048 | 0.1946 | No | ||
| 59 | SFTPC | 21759 | 11490 | -0.048 | 0.1932 | No | ||
| 60 | DBC1 | 7147 | 11589 | -0.051 | 0.1888 | No | ||
| 61 | PAX8 | 2791 14671 | 11761 | -0.055 | 0.1804 | No | ||
| 62 | CYP26B1 | 17093 | 12368 | -0.072 | 0.1489 | No | ||
| 63 | PIGN | 13865 | 12408 | -0.073 | 0.1480 | No | ||
| 64 | NTRK1 | 15299 | 12535 | -0.078 | 0.1425 | No | ||
| 65 | BTBD3 | 10457 2975 6007 6006 | 12842 | -0.088 | 0.1274 | No | ||
| 66 | EEF1A2 | 8880 14309 | 12876 | -0.090 | 0.1271 | No | ||
| 67 | MGAT5B | 20584 11197 | 12962 | -0.094 | 0.1240 | No | ||
| 68 | RTN4RL2 | 14544 | 13239 | -0.107 | 0.1109 | No | ||
| 69 | ERG | 1686 8915 | 13304 | -0.110 | 0.1092 | No | ||
| 70 | KLF15 | 17359 | 13505 | -0.123 | 0.1004 | No | ||
| 71 | BHLHB3 | 16935 | 13507 | -0.123 | 0.1024 | No | ||
| 72 | KCNS1 | 14358 | 13736 | -0.140 | 0.0924 | No | ||
| 73 | HOXB2 | 8346 | 13962 | -0.161 | 0.0829 | No | ||
| 74 | MAG | 17880 | 14107 | -0.174 | 0.0780 | No | ||
| 75 | PFN2 | 15337 1818 | 14117 | -0.175 | 0.0803 | No | ||
| 76 | IL15 | 18826 3801 | 14215 | -0.187 | 0.0782 | No | ||
| 77 | CSMD3 | 10735 7938 | 14234 | -0.189 | 0.0803 | No | ||
| 78 | STAG2 | 5521 | 14304 | -0.198 | 0.0798 | No | ||
| 79 | KCNA1 | 9202 | 14465 | -0.220 | 0.0748 | No | ||
| 80 | CREM | 2005 1965 23496 1951 | 14953 | -0.306 | 0.0535 | No | ||
| 81 | TLE4 | 23720 3697 | 15122 | -0.354 | 0.0502 | No | ||
| 82 | SERPINI1 | 340 | 15147 | -0.363 | 0.0549 | No | ||
| 83 | NR2F6 | 4651 8876 18848 8912 | 15344 | -0.425 | 0.0512 | No | ||
| 84 | CIB3 | 18840 | 15765 | -0.613 | 0.0386 | No | ||
| 85 | BRUNOL4 | 2010 4229 | 16428 | -1.064 | 0.0203 | No | ||
| 86 | TRIB2 | 21102 | 17728 | -2.918 | -0.0020 | No | ||
| 87 | GART | 22543 1754 | 17782 | -3.046 | 0.0450 | No |