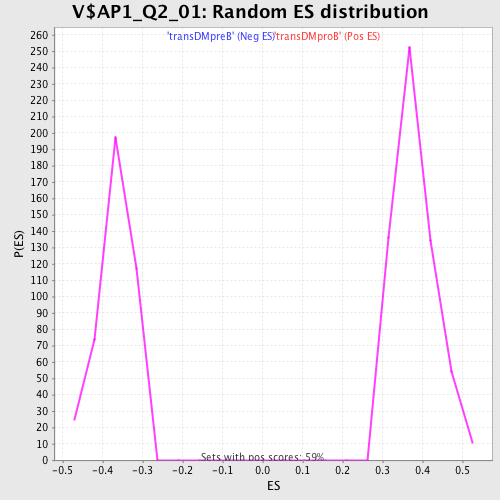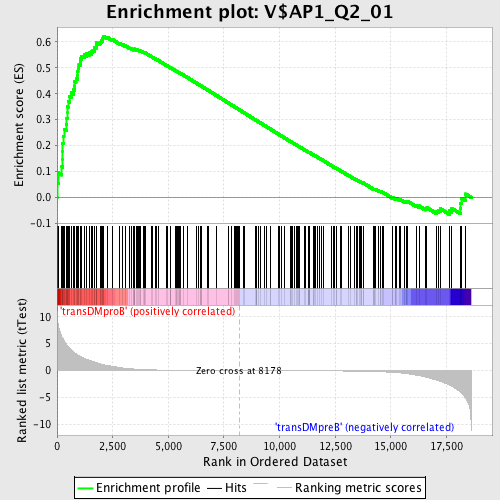
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$AP1_Q2_01 |
| Enrichment Score (ES) | 0.62218964 |
| Normalized Enrichment Score (NES) | 1.6388004 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.056608196 |
| FWER p-Value | 0.137 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IMPDH1 | 17197 1131 | 3 | 10.854 | 0.0552 | Yes | ||
| 2 | PPP1R9B | 20698 | 54 | 8.358 | 0.0951 | Yes | ||
| 3 | SQSTM1 | 9517 | 213 | 6.243 | 0.1184 | Yes | ||
| 4 | SYTL1 | 15728 | 242 | 6.022 | 0.1476 | Yes | ||
| 5 | CLIC1 | 23262 | 246 | 6.006 | 0.1781 | Yes | ||
| 6 | EDG5 | 19216 | 250 | 5.990 | 0.2085 | Yes | ||
| 7 | ATP1B1 | 4420 | 283 | 5.778 | 0.2362 | Yes | ||
| 8 | HSPB8 | 16405 | 310 | 5.542 | 0.2631 | Yes | ||
| 9 | NDEL1 | 20399 | 420 | 4.816 | 0.2817 | Yes | ||
| 10 | PKP3 | 18016 | 439 | 4.713 | 0.3048 | Yes | ||
| 11 | RIT1 | 5382 | 468 | 4.568 | 0.3266 | Yes | ||
| 12 | SNX10 | 17445 | 472 | 4.546 | 0.3496 | Yes | ||
| 13 | KRTCAP2 | 15541 | 494 | 4.444 | 0.3711 | Yes | ||
| 14 | TCF4 | 5642 | 562 | 4.128 | 0.3886 | Yes | ||
| 15 | H3F3B | 4839 | 632 | 3.843 | 0.4044 | Yes | ||
| 16 | RAP2C | 24156 | 751 | 3.382 | 0.4153 | Yes | ||
| 17 | ICK | 7135 12146 | 789 | 3.265 | 0.4299 | Yes | ||
| 18 | WASF2 | 6326 | 794 | 3.260 | 0.4463 | Yes | ||
| 19 | HCLS1 | 22770 | 850 | 3.123 | 0.4593 | Yes | ||
| 20 | AKT3 | 13739 982 | 908 | 2.985 | 0.4714 | Yes | ||
| 21 | CLDN4 | 8747 | 915 | 2.975 | 0.4863 | Yes | ||
| 22 | CDKN1A | 4511 8729 | 940 | 2.886 | 0.4997 | Yes | ||
| 23 | ITPKC | 17919 | 981 | 2.791 | 0.5118 | Yes | ||
| 24 | MECP2 | 5088 | 1047 | 2.629 | 0.5216 | Yes | ||
| 25 | CMAS | 17245 | 1059 | 2.591 | 0.5343 | Yes | ||
| 26 | LUC7L | 47 1520 | 1101 | 2.500 | 0.5448 | Yes | ||
| 27 | SLC9A1 | 16053 | 1223 | 2.254 | 0.5497 | Yes | ||
| 28 | LAMB3 | 14007 | 1331 | 2.074 | 0.5545 | Yes | ||
| 29 | PPP2CA | 20890 | 1438 | 1.912 | 0.5585 | Yes | ||
| 30 | TCF12 | 10044 3125 | 1547 | 1.753 | 0.5616 | Yes | ||
| 31 | BRD2 | 4735 8984 7628 | 1610 | 1.675 | 0.5667 | Yes | ||
| 32 | ADAM15 | 15275 | 1685 | 1.585 | 0.5708 | Yes | ||
| 33 | NR1D1 | 4650 20258 949 | 1695 | 1.572 | 0.5783 | Yes | ||
| 34 | KLF5 | 4456 | 1751 | 1.491 | 0.5830 | Yes | ||
| 35 | FHOD1 | 18772 | 1761 | 1.477 | 0.5900 | Yes | ||
| 36 | TMEM24 | 19150 | 1776 | 1.451 | 0.5966 | Yes | ||
| 37 | RBPMS | 5369 | 1928 | 1.258 | 0.5949 | Yes | ||
| 38 | ARPC5L | 15017 | 1962 | 1.211 | 0.5993 | Yes | ||
| 39 | MGST3 | 13771 12349 | 1991 | 1.179 | 0.6037 | Yes | ||
| 40 | EFNA1 | 15279 | 2031 | 1.136 | 0.6074 | Yes | ||
| 41 | AXIN2 | 20618 | 2044 | 1.118 | 0.6125 | Yes | ||
| 42 | USP3 | 19074 | 2065 | 1.103 | 0.6170 | Yes | ||
| 43 | DGKA | 3359 19589 | 2074 | 1.100 | 0.6222 | Yes | ||
| 44 | TRAPPC3 | 6553 16080 2469 11305 | 2265 | 0.970 | 0.6168 | No | ||
| 45 | COL27A1 | 6885 | 2479 | 0.793 | 0.6093 | No | ||
| 46 | SRGAP2 | 13836 17045 6433 17046 | 2812 | 0.549 | 0.5941 | No | ||
| 47 | EPHB2 | 4675 2440 8910 | 2951 | 0.476 | 0.5890 | No | ||
| 48 | ABCA2 | 15083 | 3064 | 0.426 | 0.5851 | No | ||
| 49 | SERTAD1 | 18326 | 3256 | 0.339 | 0.5765 | No | ||
| 50 | RPS20 | 7438 | 3325 | 0.312 | 0.5744 | No | ||
| 51 | LIMK2 | 9278 1296 | 3436 | 0.277 | 0.5698 | No | ||
| 52 | OBFC1 | 23648 | 3455 | 0.271 | 0.5702 | No | ||
| 53 | SLC26A9 | 11560 980 4067 | 3470 | 0.268 | 0.5708 | No | ||
| 54 | LAMA3 | 23620 2009 | 3483 | 0.265 | 0.5715 | No | ||
| 55 | ALS2CR2 | 5966 969 | 3487 | 0.264 | 0.5727 | No | ||
| 56 | NCDN | 15756 2487 6471 | 3588 | 0.236 | 0.5685 | No | ||
| 57 | GJB3 | 15754 | 3590 | 0.236 | 0.5696 | No | ||
| 58 | SYP | 9958 | 3663 | 0.219 | 0.5668 | No | ||
| 59 | ANK3 | 8591 3445 3304 3381 | 3717 | 0.207 | 0.5650 | No | ||
| 60 | GGN | 2472 1171 18312 | 3734 | 0.204 | 0.5652 | No | ||
| 61 | PSMD11 | 12772 7600 | 3763 | 0.197 | 0.5647 | No | ||
| 62 | LTBP3 | 5008 23785 9288 | 3890 | 0.171 | 0.5587 | No | ||
| 63 | EYA1 | 4695 4061 | 3923 | 0.165 | 0.5578 | No | ||
| 64 | DNAJA4 | 19446 | 3928 | 0.164 | 0.5584 | No | ||
| 65 | GAB2 | 1821 18184 2025 | 3956 | 0.160 | 0.5578 | No | ||
| 66 | ELAVL4 | 15805 4889 9137 | 4250 | 0.116 | 0.5425 | No | ||
| 67 | CLDN15 | 16667 3572 | 4304 | 0.110 | 0.5401 | No | ||
| 68 | UBE2R2 | 16240 | 4421 | 0.099 | 0.5343 | No | ||
| 69 | DNAH5 | 8470 | 4485 | 0.093 | 0.5314 | No | ||
| 70 | NCOA6 | 2876 14382 | 4570 | 0.087 | 0.5273 | No | ||
| 71 | IL11 | 17981 | 4900 | 0.068 | 0.5098 | No | ||
| 72 | CNTF | 3741 8761 3680 3736 | 4957 | 0.065 | 0.5071 | No | ||
| 73 | AQP5 | 22365 | 5098 | 0.060 | 0.4998 | No | ||
| 74 | TRIM46 | 1779 15281 | 5111 | 0.059 | 0.4994 | No | ||
| 75 | DDR2 | 13767 | 5118 | 0.059 | 0.4994 | No | ||
| 76 | TAC1 | 17526 19853 | 5302 | 0.053 | 0.4897 | No | ||
| 77 | KBTBD5 | 13020 | 5347 | 0.051 | 0.4876 | No | ||
| 78 | MITF | 17349 | 5351 | 0.051 | 0.4877 | No | ||
| 79 | LAMC2 | 4983 9266 13806 3975 | 5404 | 0.049 | 0.4851 | No | ||
| 80 | HOMER2 | 11184 | 5465 | 0.048 | 0.4821 | No | ||
| 81 | NLGN3 | 10878 | 5478 | 0.047 | 0.4817 | No | ||
| 82 | ADORA2A | 8557 | 5510 | 0.046 | 0.4803 | No | ||
| 83 | SLC41A1 | 8269 | 5542 | 0.045 | 0.4788 | No | ||
| 84 | PADI3 | 9546 | 5554 | 0.045 | 0.4784 | No | ||
| 85 | MPV17 | 9407 3505 3543 | 5663 | 0.042 | 0.4728 | No | ||
| 86 | VIT | 23155 | 5856 | 0.037 | 0.4625 | No | ||
| 87 | GFAP | 4774 | 6242 | 0.029 | 0.4418 | No | ||
| 88 | SLC6A5 | 18230 | 6344 | 0.027 | 0.4364 | No | ||
| 89 | PLCD1 | 18972 | 6429 | 0.026 | 0.4320 | No | ||
| 90 | IBRDC2 | 5747 | 6458 | 0.025 | 0.4306 | No | ||
| 91 | ELK3 | 4663 94 3388 | 6499 | 0.025 | 0.4286 | No | ||
| 92 | PTPRH | 11861 | 6504 | 0.024 | 0.4285 | No | ||
| 93 | P2RXL1 | 5218 9521 | 6769 | 0.020 | 0.4143 | No | ||
| 94 | FOXA1 | 21060 | 6817 | 0.019 | 0.4118 | No | ||
| 95 | KCNH2 | 16592 | 7166 | 0.014 | 0.3930 | No | ||
| 96 | RAB34 | 20763 | 7713 | 0.007 | 0.3634 | No | ||
| 97 | SLC30A3 | 5995 | 7719 | 0.006 | 0.3631 | No | ||
| 98 | ENO2 | 8904 | 7843 | 0.004 | 0.3565 | No | ||
| 99 | CHRM1 | 23943 | 7972 | 0.003 | 0.3495 | No | ||
| 100 | SFXN5 | 17090 | 8029 | 0.002 | 0.3465 | No | ||
| 101 | SH3RF2 | 23571 | 8049 | 0.002 | 0.3455 | No | ||
| 102 | LMOD3 | 17059 | 8123 | 0.001 | 0.3415 | No | ||
| 103 | SNCB | 21456 | 8141 | 0.000 | 0.3406 | No | ||
| 104 | DUSP9 | 24307 | 8206 | -0.000 | 0.3371 | No | ||
| 105 | MRGPRF | 17995 | 8366 | -0.003 | 0.3285 | No | ||
| 106 | DIRAS1 | 9915 | 8416 | -0.003 | 0.3259 | No | ||
| 107 | GPR3 | 15729 | 8922 | -0.009 | 0.2985 | No | ||
| 108 | UCN2 | 10308 | 8965 | -0.010 | 0.2963 | No | ||
| 109 | USP13 | 13047 | 8972 | -0.010 | 0.2960 | No | ||
| 110 | RB1CC1 | 4486 14299 | 8978 | -0.010 | 0.2958 | No | ||
| 111 | IL9 | 21444 | 9031 | -0.011 | 0.2930 | No | ||
| 112 | DYSF | 6510 | 9124 | -0.012 | 0.2881 | No | ||
| 113 | ADAMTSL1 | 16181 | 9336 | -0.015 | 0.2767 | No | ||
| 114 | PADI4 | 2424 9547 | 9427 | -0.016 | 0.2719 | No | ||
| 115 | NR0B2 | 16050 | 9572 | -0.017 | 0.2642 | No | ||
| 116 | IL1RN | 15096 | 9575 | -0.017 | 0.2641 | No | ||
| 117 | OLR1 | 8427 4233 | 9944 | -0.022 | 0.2443 | No | ||
| 118 | STARD13 | 10833 | 9997 | -0.023 | 0.2416 | No | ||
| 119 | RNF149 | 12570 | 10088 | -0.024 | 0.2368 | No | ||
| 120 | SRC | 5507 | 10226 | -0.026 | 0.2295 | No | ||
| 121 | NFATC4 | 22002 | 10479 | -0.030 | 0.2160 | No | ||
| 122 | HCN3 | 4843 1763 | 10499 | -0.030 | 0.2151 | No | ||
| 123 | RIN1 | 10349 | 10552 | -0.031 | 0.2124 | No | ||
| 124 | CXCL14 | 21445 7165 | 10580 | -0.032 | 0.2111 | No | ||
| 125 | ELAVL2 | 15839 | 10648 | -0.033 | 0.2077 | No | ||
| 126 | EIF4E | 15403 1827 8890 | 10781 | -0.035 | 0.2007 | No | ||
| 127 | MMP12 | 5108 | 10790 | -0.035 | 0.2004 | No | ||
| 128 | PTP4A1 | 9658 | 10864 | -0.036 | 0.1966 | No | ||
| 129 | FLNC | 4302 8510 | 10880 | -0.037 | 0.1960 | No | ||
| 130 | FBXO24 | 12918 3649 | 10892 | -0.037 | 0.1956 | No | ||
| 131 | AKAP1 | 4364 1326 1286 20299 | 11108 | -0.041 | 0.1841 | No | ||
| 132 | HS3ST3B1 | 7054 | 11149 | -0.042 | 0.1822 | No | ||
| 133 | COL1A2 | 8772 | 11285 | -0.044 | 0.1751 | No | ||
| 134 | LPP | 5570 | 11295 | -0.044 | 0.1748 | No | ||
| 135 | PLA2G2E | 16016 | 11297 | -0.045 | 0.1750 | No | ||
| 136 | SEMA3B | 5422 | 11322 | -0.045 | 0.1739 | No | ||
| 137 | SLC6A14 | 12168 | 11337 | -0.046 | 0.1734 | No | ||
| 138 | WNT6 | 5881 | 11353 | -0.046 | 0.1728 | No | ||
| 139 | UBE3A | 1209 1084 1830 | 11503 | -0.049 | 0.1650 | No | ||
| 140 | CRYGS | 22628 | 11509 | -0.049 | 0.1649 | No | ||
| 141 | DBC1 | 7147 | 11589 | -0.051 | 0.1609 | No | ||
| 142 | WDFY3 | 7789 3607 16462 | 11628 | -0.052 | 0.1591 | No | ||
| 143 | COBL | 1439 4539 | 11696 | -0.053 | 0.1557 | No | ||
| 144 | NTN4 | 19903 | 11784 | -0.055 | 0.1513 | No | ||
| 145 | CMYA1 | 18966 | 11881 | -0.058 | 0.1464 | No | ||
| 146 | FOSL1 | 23779 | 11984 | -0.060 | 0.1412 | No | ||
| 147 | EN1 | 387 14155 | 12319 | -0.070 | 0.1234 | No | ||
| 148 | SLITRK6 | 21724 | 12432 | -0.073 | 0.1177 | No | ||
| 149 | FAP | 14577 | 12450 | -0.074 | 0.1171 | No | ||
| 150 | SCOC | 3821 12106 | 12561 | -0.078 | 0.1116 | No | ||
| 151 | RIMS1 | 8575 | 12570 | -0.078 | 0.1115 | No | ||
| 152 | PERP | 2259 22121 20085 | 12758 | -0.085 | 0.1018 | No | ||
| 153 | ADCY8 | 22281 | 12773 | -0.085 | 0.1015 | No | ||
| 154 | TNXB | 1557 8174 878 8175 | 13110 | -0.100 | 0.0837 | No | ||
| 155 | KLF12 | 4960 2637 21732 9228 | 13177 | -0.104 | 0.0807 | No | ||
| 156 | FLI1 | 4729 | 13370 | -0.113 | 0.0708 | No | ||
| 157 | MYBPH | 14130 | 13434 | -0.118 | 0.0680 | No | ||
| 158 | VASP | 5847 | 13478 | -0.121 | 0.0663 | No | ||
| 159 | KLF15 | 17359 | 13505 | -0.123 | 0.0655 | No | ||
| 160 | PI15 | 13586 | 13588 | -0.128 | 0.0617 | No | ||
| 161 | LMNA | 4999 15290 1778 9280 | 13644 | -0.133 | 0.0594 | No | ||
| 162 | RTN4 | 7569 1328 1343 1442 | 13685 | -0.136 | 0.0579 | No | ||
| 163 | FOXL2 | 19345 | 13702 | -0.138 | 0.0578 | No | ||
| 164 | OMG | 5209 | 13750 | -0.141 | 0.0559 | No | ||
| 165 | GPR87 | 15333 | 13764 | -0.143 | 0.0560 | No | ||
| 166 | MAST2 | 9423 | 14219 | -0.187 | 0.0323 | No | ||
| 167 | MYOZ2 | 15174 | 14235 | -0.189 | 0.0324 | No | ||
| 168 | LRCH4 | 10544 6090 | 14274 | -0.194 | 0.0314 | No | ||
| 169 | NEBL | 14678 2691 | 14285 | -0.196 | 0.0318 | No | ||
| 170 | DUSP14 | 12124 | 14321 | -0.199 | 0.0309 | No | ||
| 171 | PCDH9 | 21736 | 14458 | -0.218 | 0.0247 | No | ||
| 172 | NDRG2 | 21843 | 14517 | -0.227 | 0.0227 | No | ||
| 173 | DYRK1A | 4649 | 14547 | -0.232 | 0.0223 | No | ||
| 174 | TRIB1 | 22467 | 14638 | -0.247 | 0.0186 | No | ||
| 175 | SHANK2 | 17998 | 14655 | -0.249 | 0.0190 | No | ||
| 176 | ADCK4 | 13364 | 15061 | -0.336 | -0.0012 | No | ||
| 177 | PCSK1N | 6624 | 15071 | -0.338 | 0.0000 | No | ||
| 178 | AGPAT4 | 23384 | 15209 | -0.379 | -0.0055 | No | ||
| 179 | TNFRSF12A | 23107 | 15226 | -0.384 | -0.0044 | No | ||
| 180 | CLSTN3 | 17006 | 15235 | -0.388 | -0.0029 | No | ||
| 181 | EEF1A1 | 38 38 8879 | 15411 | -0.444 | -0.0101 | No | ||
| 182 | PTPRN | 13914 | 15415 | -0.446 | -0.0080 | No | ||
| 183 | MORF4L2 | 12118 | 15611 | -0.530 | -0.0159 | No | ||
| 184 | PSME4 | 8339 20934 | 15682 | -0.567 | -0.0168 | No | ||
| 185 | TUBB4 | 22917 | 15715 | -0.586 | -0.0155 | No | ||
| 186 | GPR158 | 149 | 15752 | -0.607 | -0.0144 | No | ||
| 187 | PSMD4 | 15251 | 16145 | -0.881 | -0.0312 | No | ||
| 188 | SLC9A8 | 2842 8058 14727 | 16267 | -0.977 | -0.0328 | No | ||
| 189 | RPL23A | 11193 | 16556 | -1.204 | -0.0422 | No | ||
| 190 | NRAS | 5191 | 16623 | -1.268 | -0.0394 | No | ||
| 191 | XPO6 | 17644 | 17064 | -1.767 | -0.0542 | No | ||
| 192 | TTC1 | 7342 | 17135 | -1.859 | -0.0486 | No | ||
| 193 | ACADVL | 20375 | 17233 | -2.011 | -0.0436 | No | ||
| 194 | METAP1 | 15154 | 17641 | -2.732 | -0.0517 | No | ||
| 195 | IRAK1 | 4916 | 17740 | -2.948 | -0.0420 | No | ||
| 196 | RHBDF1 | 20504 | 18125 | -4.041 | -0.0422 | No | ||
| 197 | XPOT | 13090 | 18140 | -4.105 | -0.0221 | No | ||
| 198 | EIF4G1 | 22818 | 18175 | -4.211 | -0.0024 | No | ||
| 199 | BAG2 | 13985 | 18369 | -5.157 | 0.0134 | No |