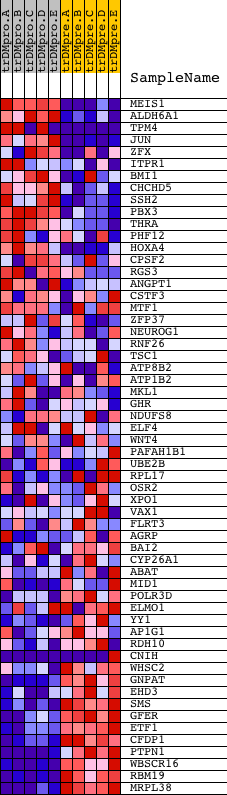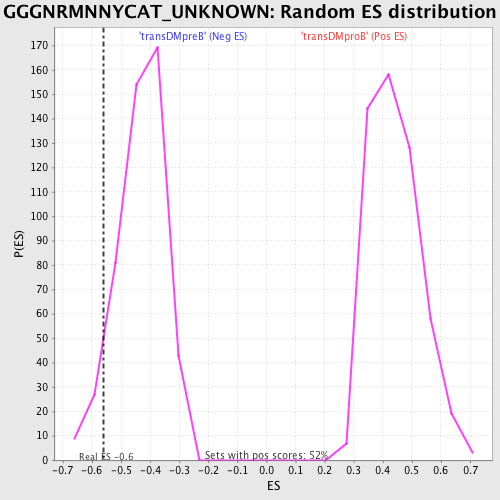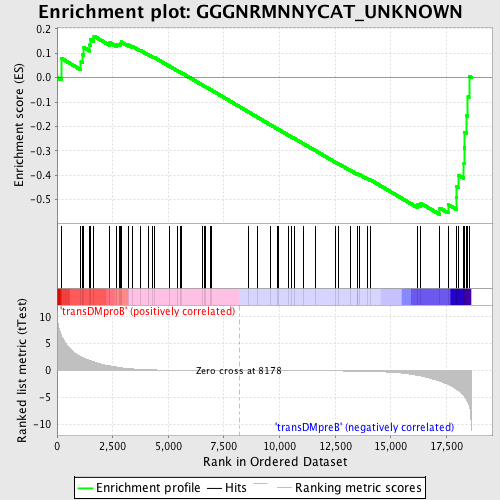
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | GGGNRMNNYCAT_UNKNOWN |
| Enrichment Score (ES) | -0.5605608 |
| Normalized Enrichment Score (NES) | -1.2975727 |
| Nominal p-value | 0.06625259 |
| FDR q-value | 0.561216 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MEIS1 | 20524 | 179 | 6.639 | 0.0797 | No | ||
| 2 | ALDH6A1 | 21025 | 1072 | 2.569 | 0.0662 | No | ||
| 3 | TPM4 | 11589 11588 6819 | 1135 | 2.448 | 0.0958 | No | ||
| 4 | JUN | 15832 | 1203 | 2.302 | 0.1232 | No | ||
| 5 | ZFX | 5984 | 1471 | 1.853 | 0.1337 | No | ||
| 6 | ITPR1 | 17341 | 1514 | 1.798 | 0.1556 | No | ||
| 7 | BMI1 | 4448 15113 | 1656 | 1.627 | 0.1699 | No | ||
| 8 | CHCHD5 | 14856 | 2369 | 0.870 | 0.1433 | No | ||
| 9 | SSH2 | 6202 | 2671 | 0.645 | 0.1357 | No | ||
| 10 | PBX3 | 9531 2714 2705 | 2784 | 0.561 | 0.1372 | No | ||
| 11 | THRA | 1447 10171 1406 | 2857 | 0.523 | 0.1404 | No | ||
| 12 | PHF12 | 11192 6483 | 2877 | 0.514 | 0.1463 | No | ||
| 13 | HOXA4 | 17148 | 3201 | 0.360 | 0.1337 | No | ||
| 14 | CPSF2 | 21182 | 3374 | 0.296 | 0.1285 | No | ||
| 15 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 3731 | 0.204 | 0.1120 | No | ||
| 16 | ANGPT1 | 8588 2260 22303 | 4099 | 0.137 | 0.0941 | No | ||
| 17 | CSTF3 | 10449 6002 | 4295 | 0.111 | 0.0851 | No | ||
| 18 | MTF1 | 5128 9421 | 4380 | 0.103 | 0.0819 | No | ||
| 19 | ZFP37 | 5962 | 5030 | 0.062 | 0.0478 | No | ||
| 20 | NEUROG1 | 21446 | 5399 | 0.050 | 0.0286 | No | ||
| 21 | RNF26 | 5614 | 5562 | 0.045 | 0.0205 | No | ||
| 22 | TSC1 | 7235 | 5570 | 0.045 | 0.0207 | No | ||
| 23 | ATP8B2 | 12055 | 6535 | 0.024 | -0.0309 | No | ||
| 24 | ATP1B2 | 20390 | 6631 | 0.023 | -0.0357 | No | ||
| 25 | MKL1 | 5864 | 6669 | 0.022 | -0.0374 | No | ||
| 26 | GHR | 22336 | 6883 | 0.019 | -0.0486 | No | ||
| 27 | NDUFS8 | 23950 | 6930 | 0.018 | -0.0509 | No | ||
| 28 | ELF4 | 24162 | 8590 | -0.005 | -0.1402 | No | ||
| 29 | WNT4 | 16025 | 8990 | -0.010 | -0.1615 | No | ||
| 30 | PAFAH1B1 | 1340 5220 9524 | 9574 | -0.017 | -0.1927 | No | ||
| 31 | UBE2B | 10245 | 9919 | -0.022 | -0.2110 | No | ||
| 32 | RPL17 | 11429 6653 | 9962 | -0.023 | -0.2129 | No | ||
| 33 | OSR2 | 22492 | 10392 | -0.029 | -0.2356 | No | ||
| 34 | XPO1 | 4172 | 10543 | -0.031 | -0.2433 | No | ||
| 35 | VAX1 | 23636 | 10663 | -0.033 | -0.2493 | No | ||
| 36 | FLRT3 | 7730 12930 2673 | 11069 | -0.040 | -0.2706 | No | ||
| 37 | AGRP | 18768 | 11595 | -0.051 | -0.2982 | No | ||
| 38 | BAI2 | 2541 2379 16067 | 12532 | -0.077 | -0.3475 | No | ||
| 39 | CYP26A1 | 23871 3695 | 12654 | -0.082 | -0.3530 | No | ||
| 40 | ABAT | 22864 1713 22669 | 13182 | -0.104 | -0.3800 | No | ||
| 41 | MID1 | 5097 5098 | 13495 | -0.122 | -0.3951 | No | ||
| 42 | POLR3D | 21760 12456 | 13499 | -0.122 | -0.3937 | No | ||
| 43 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13606 | -0.130 | -0.3976 | No | ||
| 44 | YY1 | 10371 | 13934 | -0.157 | -0.4131 | No | ||
| 45 | AP1G1 | 4392 | 14089 | -0.173 | -0.4191 | No | ||
| 46 | RDH10 | 8272 | 16189 | -0.914 | -0.5199 | No | ||
| 47 | CNIH | 4536 23971 | 16348 | -1.015 | -0.5148 | No | ||
| 48 | WHSC2 | 6298 | 17199 | -1.962 | -0.5342 | Yes | ||
| 49 | GNPAT | 18420 | 17576 | -2.579 | -0.5197 | Yes | ||
| 50 | EHD3 | 23159 | 17941 | -3.473 | -0.4926 | Yes | ||
| 51 | SMS | 2573 24023 | 17949 | -3.495 | -0.4460 | Yes | ||
| 52 | GFER | 23088 | 18051 | -3.785 | -0.4005 | Yes | ||
| 53 | ETF1 | 23467 | 18287 | -4.669 | -0.3503 | Yes | ||
| 54 | CFDP1 | 10710 | 18294 | -4.702 | -0.2874 | Yes | ||
| 55 | PTPN1 | 5325 | 18317 | -4.853 | -0.2233 | Yes | ||
| 56 | WBSCR16 | 16355 | 18411 | -5.418 | -0.1554 | Yes | ||
| 57 | RBM19 | 13164 | 18467 | -5.974 | -0.0780 | Yes | ||
| 58 | MRPL38 | 20145 | 18515 | -6.388 | 0.0054 | Yes |