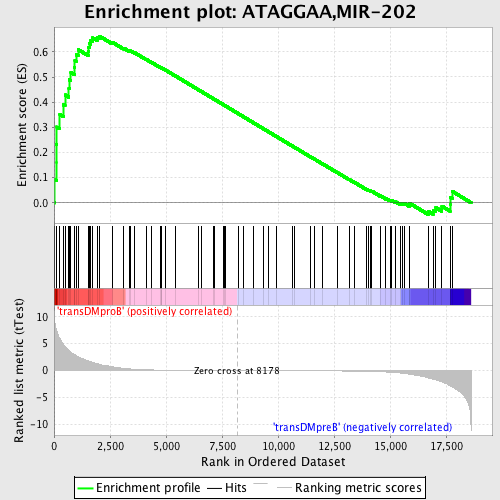
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | ATAGGAA,MIR-202 |
| Enrichment Score (ES) | 0.66254956 |
| Normalized Enrichment Score (NES) | 1.5722446 |
| Nominal p-value | 0.0018416207 |
| FDR q-value | 0.049435563 |
| FWER p-Value | 0.423 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MARCKS | 9331 | 19 | 9.740 | 0.0927 | Yes | ||
| 2 | ZFAND3 | 23306 | 110 | 7.470 | 0.1598 | Yes | ||
| 3 | NDRG1 | 22276 | 112 | 7.468 | 0.2316 | Yes | ||
| 4 | CNN3 | 12983 | 125 | 7.309 | 0.3013 | Yes | ||
| 5 | ELF1 | 21948 | 256 | 5.939 | 0.3515 | Yes | ||
| 6 | BCL11A | 4691 | 412 | 4.861 | 0.3899 | Yes | ||
| 7 | RNF11 | 11383 2509 | 490 | 4.470 | 0.4288 | Yes | ||
| 8 | PTEN | 5305 | 661 | 3.694 | 0.4551 | Yes | ||
| 9 | STAT3 | 5525 9906 | 681 | 3.642 | 0.4892 | Yes | ||
| 10 | LBR | 13732 | 749 | 3.387 | 0.5182 | Yes | ||
| 11 | ATXN1 | 21477 | 918 | 2.967 | 0.5376 | Yes | ||
| 12 | RASA1 | 10174 | 929 | 2.929 | 0.5653 | Yes | ||
| 13 | KRAS | 9247 | 987 | 2.779 | 0.5890 | Yes | ||
| 14 | ACVR1 | 4334 | 1076 | 2.564 | 0.6089 | Yes | ||
| 15 | RAD52 | 17306 | 1521 | 1.786 | 0.6022 | Yes | ||
| 16 | TCF12 | 10044 3125 | 1547 | 1.753 | 0.6177 | Yes | ||
| 17 | THRAP2 | 8009 | 1570 | 1.723 | 0.6331 | Yes | ||
| 18 | SSBP2 | 3171 7364 3252 | 1637 | 1.635 | 0.6453 | Yes | ||
| 19 | CREBBP | 22682 8783 | 1722 | 1.533 | 0.6555 | Yes | ||
| 20 | MAPK6 | 19062 | 1923 | 1.265 | 0.6569 | Yes | ||
| 21 | ANP32E | 15500 | 2023 | 1.146 | 0.6625 | Yes | ||
| 22 | MIB1 | 5908 | 2605 | 0.692 | 0.6379 | No | ||
| 23 | USP15 | 4758 3377 | 3102 | 0.407 | 0.6150 | No | ||
| 24 | HIP2 | 3626 3485 16838 | 3360 | 0.301 | 0.6041 | No | ||
| 25 | APPBP2 | 7357 | 3416 | 0.284 | 0.6039 | No | ||
| 26 | GRIA3 | 24349 11996 2649 | 3595 | 0.235 | 0.5965 | No | ||
| 27 | CTBP2 | 17591 1137 | 4132 | 0.132 | 0.5689 | No | ||
| 28 | TRIM33 | 8236 13579 15472 8237 | 4347 | 0.106 | 0.5584 | No | ||
| 29 | CD28 | 14239 4092 | 4745 | 0.076 | 0.5377 | No | ||
| 30 | EAF1 | 3634 22055 | 4785 | 0.073 | 0.5363 | No | ||
| 31 | PCGF2 | 20270 | 4794 | 0.073 | 0.5365 | No | ||
| 32 | VANGL2 | 4109 8220 | 4978 | 0.064 | 0.5273 | No | ||
| 33 | EI24 | 19179 | 5434 | 0.048 | 0.5032 | No | ||
| 34 | TGFBR2 | 2994 3001 10167 | 6456 | 0.025 | 0.4484 | No | ||
| 35 | RAB22A | 2888 5343 | 6573 | 0.023 | 0.4424 | No | ||
| 36 | HLF | 10124 | 7107 | 0.015 | 0.4138 | No | ||
| 37 | ARSB | 4415 439 | 7158 | 0.014 | 0.4112 | No | ||
| 38 | LUZP1 | 6533 11257 2503 | 7571 | 0.009 | 0.3891 | No | ||
| 39 | RNF121 | 7952 373 | 7616 | 0.008 | 0.3868 | No | ||
| 40 | ACSL3 | 13184 | 7629 | 0.008 | 0.3862 | No | ||
| 41 | DDEF1 | 22282 2253 | 8230 | -0.001 | 0.3539 | No | ||
| 42 | SNAP91 | 9839 19043 | 8439 | -0.004 | 0.3427 | No | ||
| 43 | KIAA1715 | 7629 | 8915 | -0.009 | 0.3171 | No | ||
| 44 | PPARBP | 1203 20263 1195 | 9335 | -0.015 | 0.2947 | No | ||
| 45 | DNAJC10 | 7349 | 9582 | -0.018 | 0.2816 | No | ||
| 46 | RPS6KA3 | 8490 | 9929 | -0.022 | 0.2631 | No | ||
| 47 | CPEB3 | 5539 | 10625 | -0.033 | 0.2260 | No | ||
| 48 | USP8 | 2757 2833 14869 | 10714 | -0.034 | 0.2216 | No | ||
| 49 | RKHD3 | 18199 | 11443 | -0.048 | 0.1827 | No | ||
| 50 | KHDRBS2 | 14284 | 11629 | -0.052 | 0.1733 | No | ||
| 51 | ESR1 | 20097 4685 | 11994 | -0.061 | 0.1542 | No | ||
| 52 | FAM60A | 12094 | 12630 | -0.081 | 0.1208 | No | ||
| 53 | KLF12 | 4960 2637 21732 9228 | 13177 | -0.104 | 0.0923 | No | ||
| 54 | ERBB2IP | 3232 12234 | 13415 | -0.117 | 0.0806 | No | ||
| 55 | HOXB2 | 8346 | 13962 | -0.161 | 0.0527 | No | ||
| 56 | NR2F2 | 8619 409 | 14034 | -0.169 | 0.0505 | No | ||
| 57 | ENAH | 4665 8901 | 14118 | -0.175 | 0.0477 | No | ||
| 58 | SORCS1 | 7184 12203 | 14184 | -0.183 | 0.0460 | No | ||
| 59 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 14565 | -0.235 | 0.0278 | No | ||
| 60 | SPRED1 | 4331 | 14814 | -0.274 | 0.0170 | No | ||
| 61 | CBL | 19154 | 15001 | -0.315 | 0.0100 | No | ||
| 62 | SFPQ | 12936 | 15076 | -0.341 | 0.0093 | No | ||
| 63 | ROBO2 | 6501 | 15223 | -0.383 | 0.0051 | No | ||
| 64 | SNX16 | 15374 | 15465 | -0.467 | -0.0034 | No | ||
| 65 | YAF2 | 22155 | 15564 | -0.512 | -0.0037 | No | ||
| 66 | IQGAP1 | 6619 | 15622 | -0.539 | -0.0016 | No | ||
| 67 | BICD2 | 8048 | 15864 | -0.666 | -0.0082 | No | ||
| 68 | BTG1 | 4458 4457 8663 | 15877 | -0.677 | -0.0023 | No | ||
| 69 | TARDBP | 10520 6066 | 16700 | -1.363 | -0.0335 | No | ||
| 70 | NARG1 | 7934 917 | 16919 | -1.628 | -0.0296 | No | ||
| 71 | SENP1 | 10301 | 17041 | -1.737 | -0.0194 | No | ||
| 72 | PPP5C | 17952 | 17311 | -2.119 | -0.0135 | No | ||
| 73 | BAG4 | 7431 | 17677 | -2.813 | -0.0062 | No | ||
| 74 | TTC13 | 10636 | 17687 | -2.839 | 0.0207 | No | ||
| 75 | DDX3X | 24379 | 17785 | -3.050 | 0.0448 | No |

