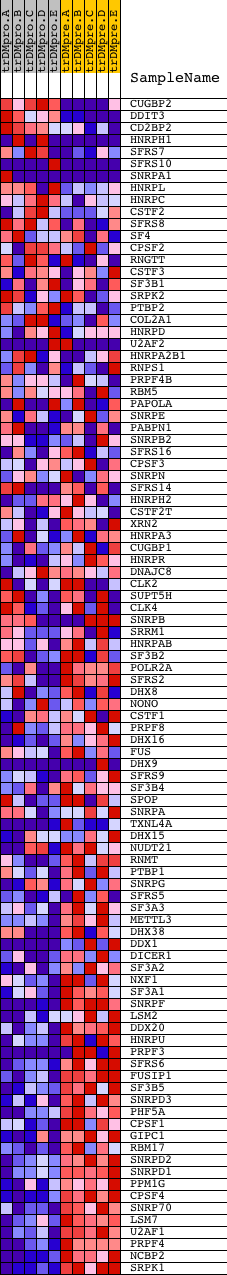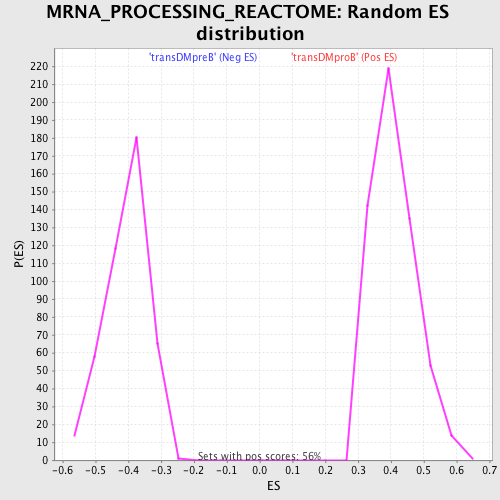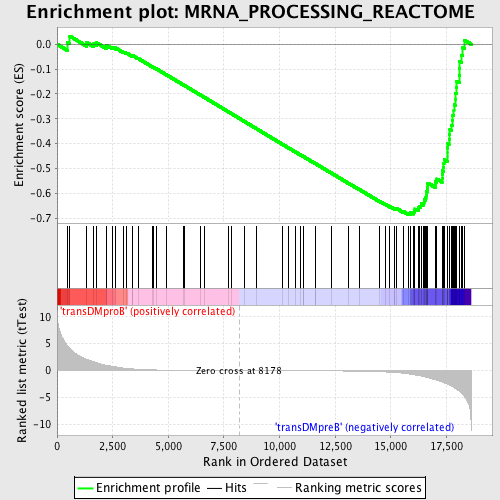
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | MRNA_PROCESSING_REACTOME |
| Enrichment Score (ES) | -0.68486303 |
| Normalized Enrichment Score (NES) | -1.6954855 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.023704665 |
| FWER p-Value | 0.068 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CUGBP2 | 4687 8923 | 457 | 4.622 | 0.0091 | No | ||
| 2 | DDIT3 | 19857 | 573 | 4.086 | 0.0328 | No | ||
| 3 | CD2BP2 | 17621 | 1337 | 2.070 | 0.0067 | No | ||
| 4 | HNRPH1 | 12222 1486 1220 7202 7201 | 1654 | 1.627 | 0.0016 | No | ||
| 5 | SFRS7 | 22889 | 1763 | 1.472 | 0.0065 | No | ||
| 6 | SFRS10 | 9818 1722 5441 | 2217 | 0.998 | -0.0106 | No | ||
| 7 | SNRPA1 | 1373 18216 | 2234 | 0.991 | -0.0043 | No | ||
| 8 | HNRPL | 18315 | 2503 | 0.778 | -0.0130 | No | ||
| 9 | HNRPC | 9102 4859 | 2620 | 0.684 | -0.0143 | No | ||
| 10 | CSTF2 | 24257 | 2997 | 0.456 | -0.0313 | No | ||
| 11 | SFRS8 | 10543 6089 | 3124 | 0.394 | -0.0352 | No | ||
| 12 | SF4 | 18593 | 3372 | 0.297 | -0.0464 | No | ||
| 13 | CPSF2 | 21182 | 3374 | 0.296 | -0.0443 | No | ||
| 14 | RNGTT | 10769 2354 6272 | 3641 | 0.224 | -0.0570 | No | ||
| 15 | CSTF3 | 10449 6002 | 4295 | 0.111 | -0.0914 | No | ||
| 16 | SF3B1 | 13954 | 4344 | 0.106 | -0.0932 | No | ||
| 17 | SRPK2 | 5513 | 4466 | 0.095 | -0.0991 | No | ||
| 18 | PTBP2 | 7074 | 4914 | 0.067 | -0.1227 | No | ||
| 19 | COL2A1 | 22145 4542 | 5665 | 0.042 | -0.1629 | No | ||
| 20 | HNRPD | 8645 | 5720 | 0.041 | -0.1655 | No | ||
| 21 | U2AF2 | 5813 5812 5814 | 6448 | 0.026 | -0.2046 | No | ||
| 22 | HNRPA2B1 | 1187 7001 7000 1040 | 6613 | 0.023 | -0.2133 | No | ||
| 23 | RNPS1 | 9730 23361 | 7695 | 0.007 | -0.2716 | No | ||
| 24 | PRPF4B | 3208 5295 | 7854 | 0.004 | -0.2801 | No | ||
| 25 | RBM5 | 13507 3016 | 8413 | -0.003 | -0.3102 | No | ||
| 26 | PAPOLA | 5261 9585 | 8954 | -0.010 | -0.3393 | No | ||
| 27 | SNRPE | 9843 | 10126 | -0.025 | -0.4024 | No | ||
| 28 | PABPN1 | 12018 | 10393 | -0.029 | -0.4165 | No | ||
| 29 | SNRPB2 | 5470 | 10695 | -0.034 | -0.4325 | No | ||
| 30 | SFRS16 | 17943 3840 | 10933 | -0.038 | -0.4451 | No | ||
| 31 | CPSF3 | 7038 | 11079 | -0.040 | -0.4526 | No | ||
| 32 | SNRPN | 1715 9844 5471 | 11092 | -0.041 | -0.4529 | No | ||
| 33 | SFRS14 | 10622 | 11617 | -0.052 | -0.4809 | No | ||
| 34 | HNRPH2 | 12085 7082 | 11631 | -0.052 | -0.4812 | No | ||
| 35 | CSTF2T | 13502 | 12311 | -0.070 | -0.5173 | No | ||
| 36 | XRN2 | 2936 6302 2905 | 13107 | -0.100 | -0.5595 | No | ||
| 37 | HNRPA3 | 6023 2712 10473 2683 | 13589 | -0.129 | -0.5846 | No | ||
| 38 | CUGBP1 | 2805 8819 4576 2924 | 14499 | -0.225 | -0.6320 | No | ||
| 39 | HNRPR | 13196 7909 2500 | 14743 | -0.263 | -0.6432 | No | ||
| 40 | DNAJC8 | 12712 | 14961 | -0.308 | -0.6527 | No | ||
| 41 | CLK2 | 1883 15544 | 15163 | -0.366 | -0.6608 | No | ||
| 42 | SUPT5H | 9939 | 15184 | -0.372 | -0.6592 | No | ||
| 43 | CLK4 | 20895 1292 | 15232 | -0.386 | -0.6589 | No | ||
| 44 | SNRPB | 9842 5469 2736 | 15571 | -0.515 | -0.6734 | No | ||
| 45 | SRRM1 | 6958 | 15782 | -0.618 | -0.6802 | Yes | ||
| 46 | HNRPAB | 9104 | 15869 | -0.673 | -0.6799 | Yes | ||
| 47 | SF3B2 | 23974 | 15888 | -0.682 | -0.6759 | Yes | ||
| 48 | POLR2A | 5394 | 16021 | -0.780 | -0.6774 | Yes | ||
| 49 | SFRS2 | 9807 20136 | 16049 | -0.798 | -0.6730 | Yes | ||
| 50 | DHX8 | 20649 | 16062 | -0.808 | -0.6677 | Yes | ||
| 51 | NONO | 11993 | 16084 | -0.831 | -0.6628 | Yes | ||
| 52 | CSTF1 | 12515 | 16233 | -0.954 | -0.6638 | Yes | ||
| 53 | PRPF8 | 20780 1371 | 16257 | -0.972 | -0.6579 | Yes | ||
| 54 | DHX16 | 23247 | 16271 | -0.980 | -0.6515 | Yes | ||
| 55 | FUS | 6135 | 16356 | -1.019 | -0.6485 | Yes | ||
| 56 | DHX9 | 8845 4608 3948 | 16367 | -1.024 | -0.6416 | Yes | ||
| 57 | SFRS9 | 16731 | 16466 | -1.109 | -0.6388 | Yes | ||
| 58 | SF3B4 | 22269 | 16492 | -1.131 | -0.6318 | Yes | ||
| 59 | SPOP | 5498 | 16515 | -1.153 | -0.6246 | Yes | ||
| 60 | SNRPA | 7007 11992 7008 | 16563 | -1.214 | -0.6183 | Yes | ||
| 61 | TXNL4A | 6567 11329 | 16582 | -1.228 | -0.6103 | Yes | ||
| 62 | DHX15 | 8842 | 16588 | -1.232 | -0.6015 | Yes | ||
| 63 | NUDT21 | 12665 | 16589 | -1.233 | -0.5925 | Yes | ||
| 64 | RNMT | 7501 | 16629 | -1.276 | -0.5853 | Yes | ||
| 65 | PTBP1 | 5303 | 16652 | -1.298 | -0.5770 | Yes | ||
| 66 | SNRPG | 12622 | 16664 | -1.317 | -0.5679 | Yes | ||
| 67 | SFRS5 | 9808 2062 | 16665 | -1.319 | -0.5583 | Yes | ||
| 68 | SF3A3 | 16091 | 17000 | -1.699 | -0.5639 | Yes | ||
| 69 | METTL3 | 21841 | 17006 | -1.704 | -0.5517 | Yes | ||
| 70 | DHX38 | 48 | 17069 | -1.776 | -0.5421 | Yes | ||
| 71 | DDX1 | 21104 | 17303 | -2.109 | -0.5392 | Yes | ||
| 72 | DICER1 | 20989 | 17314 | -2.121 | -0.5242 | Yes | ||
| 73 | SF3A2 | 19938 | 17326 | -2.138 | -0.5092 | Yes | ||
| 74 | NXF1 | 11986 | 17370 | -2.218 | -0.4953 | Yes | ||
| 75 | SF3A1 | 7450 | 17384 | -2.247 | -0.4796 | Yes | ||
| 76 | SNRPF | 7645 | 17393 | -2.259 | -0.4635 | Yes | ||
| 77 | LSM2 | 23266 | 17543 | -2.501 | -0.4532 | Yes | ||
| 78 | DDX20 | 15213 | 17544 | -2.502 | -0.4349 | Yes | ||
| 79 | HNRPU | 6959 | 17546 | -2.506 | -0.4166 | Yes | ||
| 80 | PRPF3 | 12899 12898 1877 7703 | 17562 | -2.549 | -0.3988 | Yes | ||
| 81 | SFRS6 | 14751 | 17617 | -2.689 | -0.3821 | Yes | ||
| 82 | FUSIP1 | 4715 16036 | 17627 | -2.708 | -0.3627 | Yes | ||
| 83 | SF3B5 | 7256 | 17655 | -2.755 | -0.3440 | Yes | ||
| 84 | SNRPD3 | 12514 | 17723 | -2.911 | -0.3264 | Yes | ||
| 85 | PHF5A | 22194 | 17755 | -2.970 | -0.3063 | Yes | ||
| 86 | CPSF1 | 22241 | 17788 | -3.055 | -0.2857 | Yes | ||
| 87 | GIPC1 | 3841 3882 18552 | 17837 | -3.154 | -0.2652 | Yes | ||
| 88 | RBM17 | 13371 | 17840 | -3.163 | -0.2422 | Yes | ||
| 89 | SNRPD2 | 8412 | 17892 | -3.333 | -0.2206 | Yes | ||
| 90 | SNRPD1 | 23622 | 17897 | -3.342 | -0.1964 | Yes | ||
| 91 | PPM1G | 16571 | 17939 | -3.465 | -0.1732 | Yes | ||
| 92 | CPSF4 | 16627 | 17967 | -3.556 | -0.1487 | Yes | ||
| 93 | SNRP70 | 1186 | 18077 | -3.843 | -0.1265 | Yes | ||
| 94 | LSM7 | 12286 | 18089 | -3.878 | -0.0987 | Yes | ||
| 95 | U2AF1 | 8431 | 18097 | -3.895 | -0.0706 | Yes | ||
| 96 | PRPF4 | 7655 12845 | 18177 | -4.216 | -0.0440 | Yes | ||
| 97 | NCBP2 | 12643 | 18234 | -4.445 | -0.0145 | Yes | ||
| 98 | SRPK1 | 23049 | 18311 | -4.799 | 0.0165 | Yes |