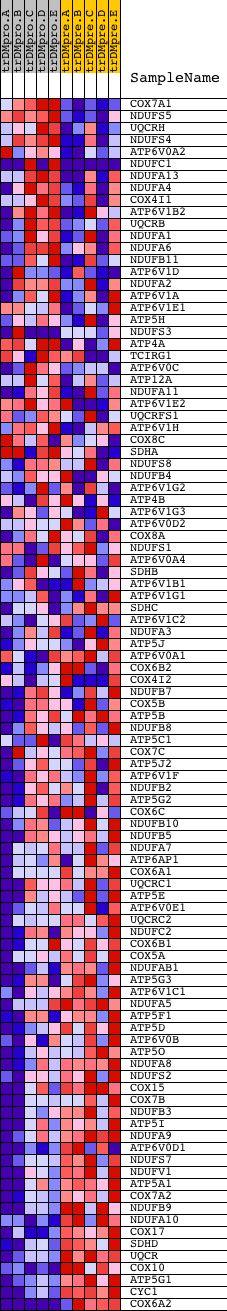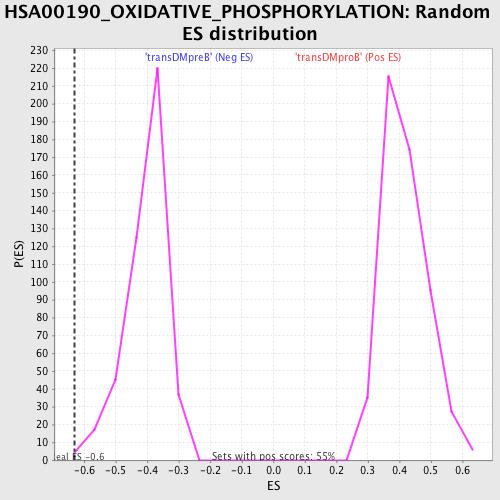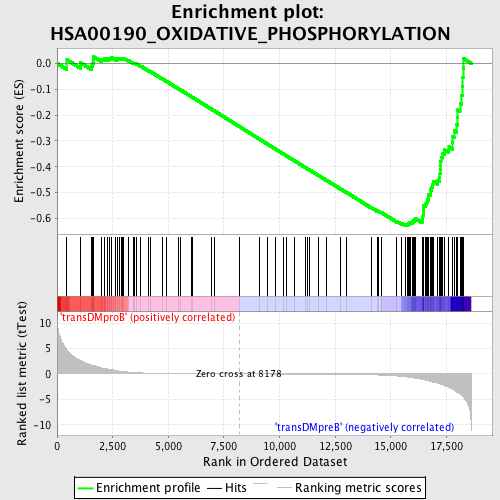
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | HSA00190_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.6290783 |
| Normalized Enrichment Score (NES) | -1.5709857 |
| Nominal p-value | 0.002232143 |
| FDR q-value | 0.08796275 |
| FWER p-Value | 0.635 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COX7A1 | 4554 | 443 | 4.681 | 0.0142 | No | ||
| 2 | NDUFS5 | 9296 | 1055 | 2.601 | 0.0024 | No | ||
| 3 | UQCRH | 12378 | 1549 | 1.750 | -0.0100 | No | ||
| 4 | NDUFS4 | 9452 5157 3173 | 1595 | 1.694 | 0.0014 | No | ||
| 5 | ATP6V0A2 | 5760 | 1623 | 1.650 | 0.0133 | No | ||
| 6 | NDUFC1 | 7287 12339 | 1648 | 1.630 | 0.0253 | No | ||
| 7 | NDUFA13 | 7400 | 1986 | 1.183 | 0.0167 | No | ||
| 8 | NDUFA4 | 9451 | 2118 | 1.049 | 0.0182 | No | ||
| 9 | COX4I1 | 18444 | 2252 | 0.981 | 0.0190 | No | ||
| 10 | ATP6V1B2 | 18599 | 2364 | 0.874 | 0.0201 | No | ||
| 11 | UQCRB | 12545 | 2453 | 0.811 | 0.0220 | No | ||
| 12 | NDUFA1 | 24170 | 2640 | 0.672 | 0.0174 | No | ||
| 13 | NDUFA6 | 12473 | 2724 | 0.611 | 0.0179 | No | ||
| 14 | NDUFB11 | 24176 | 2792 | 0.558 | 0.0188 | No | ||
| 15 | ATP6V1D | 7872 | 2880 | 0.513 | 0.0183 | No | ||
| 16 | NDUFA2 | 9450 | 2943 | 0.478 | 0.0188 | No | ||
| 17 | ATP6V1A | 8638 | 3005 | 0.453 | 0.0192 | No | ||
| 18 | ATP6V1E1 | 4423 | 3229 | 0.348 | 0.0100 | No | ||
| 19 | ATP5H | 12948 | 3454 | 0.271 | 0.0001 | No | ||
| 20 | NDUFS3 | 7553 12683 | 3498 | 0.260 | -0.0001 | No | ||
| 21 | ATP4A | 4422 | 3571 | 0.240 | -0.0020 | No | ||
| 22 | TCIRG1 | 3700 3752 23951 | 3757 | 0.198 | -0.0104 | No | ||
| 23 | ATP6V0C | 8643 | 4124 | 0.133 | -0.0291 | No | ||
| 24 | ATP12A | 16700 | 4204 | 0.121 | -0.0323 | No | ||
| 25 | NDUFA11 | 12832 | 4731 | 0.077 | -0.0601 | No | ||
| 26 | ATP6V1E2 | 22880 | 4933 | 0.066 | -0.0704 | No | ||
| 27 | UQCRFS1 | 21499 | 5439 | 0.048 | -0.0973 | No | ||
| 28 | ATP6V1H | 14301 | 5552 | 0.045 | -0.1030 | No | ||
| 29 | COX8C | 21175 | 6019 | 0.034 | -0.1279 | No | ||
| 30 | SDHA | 12436 | 6065 | 0.033 | -0.1301 | No | ||
| 31 | NDUFS8 | 23950 | 6930 | 0.018 | -0.1766 | No | ||
| 32 | NDUFB4 | 22596 7541 | 7075 | 0.015 | -0.1842 | No | ||
| 33 | ATP6V1G2 | 23257 1534 | 8192 | -0.000 | -0.2445 | No | ||
| 34 | ATP4B | 18928 | 9103 | -0.012 | -0.2935 | No | ||
| 35 | ATP6V1G3 | 14107 | 9445 | -0.016 | -0.3118 | No | ||
| 36 | ATP6V0D2 | 15934 | 9805 | -0.021 | -0.3311 | No | ||
| 37 | COX8A | 8777 | 10171 | -0.026 | -0.3506 | No | ||
| 38 | NDUFS1 | 13940 | 10315 | -0.028 | -0.3581 | No | ||
| 39 | ATP6V0A4 | 8934 | 10662 | -0.033 | -0.3765 | No | ||
| 40 | SDHB | 2348 12566 14880 | 11148 | -0.042 | -0.4023 | No | ||
| 41 | ATP6V1B1 | 8499 | 11276 | -0.044 | -0.4088 | No | ||
| 42 | ATP6V1G1 | 12322 | 11350 | -0.046 | -0.4124 | No | ||
| 43 | SDHC | 7248 12279 | 11770 | -0.055 | -0.4346 | No | ||
| 44 | ATP6V1C2 | 21098 | 12094 | -0.063 | -0.4515 | No | ||
| 45 | NDUFA3 | 18410 | 12750 | -0.085 | -0.4862 | No | ||
| 46 | ATP5J | 880 22555 | 13015 | -0.095 | -0.4997 | No | ||
| 47 | ATP6V0A1 | 8640 4424 1197 | 14114 | -0.175 | -0.5575 | No | ||
| 48 | COX6B2 | 17983 4110 | 14389 | -0.208 | -0.5707 | No | ||
| 49 | COX4I2 | 14791 | 14439 | -0.216 | -0.5715 | No | ||
| 50 | NDUFB7 | 12429 | 14567 | -0.236 | -0.5765 | No | ||
| 51 | COX5B | 4552 | 15275 | -0.402 | -0.6114 | No | ||
| 52 | ATP5B | 19846 | 15495 | -0.481 | -0.6193 | No | ||
| 53 | NDUFB8 | 7418 | 15677 | -0.562 | -0.6245 | Yes | ||
| 54 | ATP5C1 | 8635 | 15742 | -0.601 | -0.6231 | Yes | ||
| 55 | COX7C | 8776 | 15785 | -0.619 | -0.6203 | Yes | ||
| 56 | ATP5J2 | 12186 | 15825 | -0.642 | -0.6172 | Yes | ||
| 57 | ATP6V1F | 12291 | 15889 | -0.683 | -0.6150 | Yes | ||
| 58 | NDUFB2 | 7542 | 15959 | -0.733 | -0.6128 | Yes | ||
| 59 | ATP5G2 | 12610 | 16008 | -0.771 | -0.6091 | Yes | ||
| 60 | COX6C | 8775 | 16060 | -0.803 | -0.6053 | Yes | ||
| 61 | NDUFB10 | 23084 | 16120 | -0.862 | -0.6015 | Yes | ||
| 62 | NDUFB5 | 12277 | 16405 | -1.046 | -0.6083 | Yes | ||
| 63 | NDUFA7 | 12347 | 16418 | -1.055 | -0.6003 | Yes | ||
| 64 | ATP6AP1 | 24296 | 16438 | -1.081 | -0.5926 | Yes | ||
| 65 | COX6A1 | 8774 4553 | 16448 | -1.093 | -0.5842 | Yes | ||
| 66 | UQCRC1 | 10259 | 16459 | -1.103 | -0.5757 | Yes | ||
| 67 | ATP5E | 14321 | 16460 | -1.105 | -0.5667 | Yes | ||
| 68 | ATP6V0E1 | 23326 | 16484 | -1.124 | -0.5588 | Yes | ||
| 69 | UQCRC2 | 18102 | 16485 | -1.125 | -0.5496 | Yes | ||
| 70 | NDUFC2 | 18183 | 16564 | -1.215 | -0.5440 | Yes | ||
| 71 | COX6B1 | 4283 8479 | 16621 | -1.266 | -0.5367 | Yes | ||
| 72 | COX5A | 19431 | 16661 | -1.310 | -0.5281 | Yes | ||
| 73 | NDUFAB1 | 7667 | 16683 | -1.340 | -0.5183 | Yes | ||
| 74 | ATP5G3 | 14558 | 16694 | -1.357 | -0.5078 | Yes | ||
| 75 | ATP6V1C1 | 22487 12329 | 16790 | -1.477 | -0.5009 | Yes | ||
| 76 | NDUFA5 | 1076 7544 | 16798 | -1.480 | -0.4893 | Yes | ||
| 77 | ATP5F1 | 15212 | 16834 | -1.531 | -0.4787 | Yes | ||
| 78 | ATP5D | 19949 | 16886 | -1.587 | -0.4685 | Yes | ||
| 79 | ATP6V0B | 15786 | 16896 | -1.602 | -0.4560 | Yes | ||
| 80 | ATP5O | 22539 | 17117 | -1.839 | -0.4529 | Yes | ||
| 81 | NDUFA8 | 14605 | 17195 | -1.957 | -0.4411 | Yes | ||
| 82 | NDUFS2 | 4089 13760 | 17208 | -1.977 | -0.4256 | Yes | ||
| 83 | COX15 | 5931 | 17225 | -1.998 | -0.4102 | Yes | ||
| 84 | COX7B | 7260 | 17234 | -2.013 | -0.3943 | Yes | ||
| 85 | NDUFB3 | 12362 | 17240 | -2.028 | -0.3780 | Yes | ||
| 86 | ATP5I | 8637 | 17258 | -2.054 | -0.3622 | Yes | ||
| 87 | NDUFA9 | 12288 | 17331 | -2.149 | -0.3486 | Yes | ||
| 88 | ATP6V0D1 | 3895 8639 3920 | 17425 | -2.305 | -0.3349 | Yes | ||
| 89 | NDUFS7 | 19946 | 17613 | -2.682 | -0.3231 | Yes | ||
| 90 | NDUFV1 | 23953 | 17773 | -3.028 | -0.3070 | Yes | ||
| 91 | ATP5A1 | 23505 | 17790 | -3.062 | -0.2830 | Yes | ||
| 92 | COX7A2 | 19053 | 17845 | -3.175 | -0.2600 | Yes | ||
| 93 | NDUFB9 | 12306 | 17969 | -3.558 | -0.2377 | Yes | ||
| 94 | NDUFA10 | 7420 | 18001 | -3.639 | -0.2097 | Yes | ||
| 95 | COX17 | 467 | 18008 | -3.679 | -0.1801 | Yes | ||
| 96 | SDHD | 19125 | 18110 | -3.948 | -0.1534 | Yes | ||
| 97 | UQCR | 12382 | 18193 | -4.275 | -0.1230 | Yes | ||
| 98 | COX10 | 20408 | 18215 | -4.368 | -0.0886 | Yes | ||
| 99 | ATP5G1 | 8636 | 18230 | -4.437 | -0.0532 | Yes | ||
| 100 | CYC1 | 12348 | 18248 | -4.488 | -0.0176 | Yes | ||
| 101 | COX6A2 | 17605 | 18270 | -4.596 | 0.0187 | Yes |