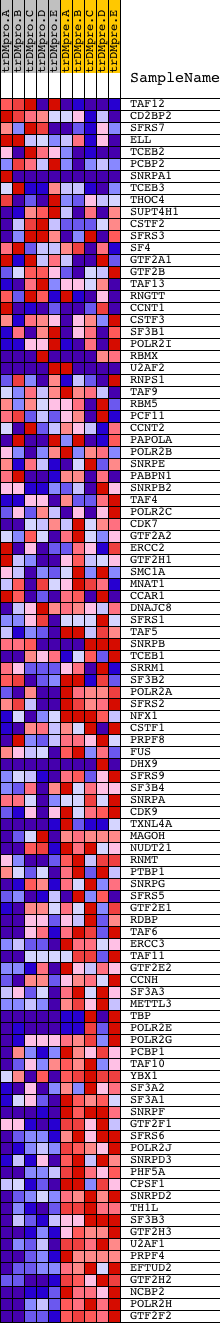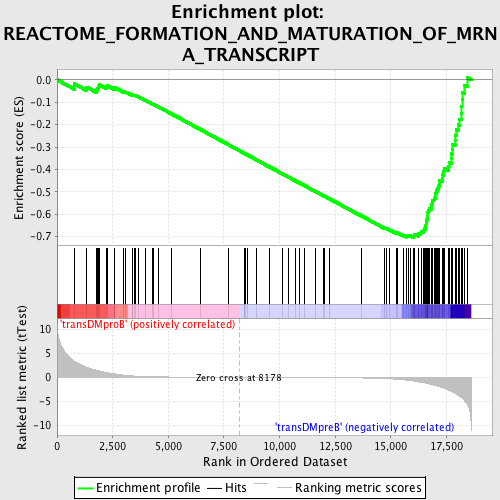
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | REACTOME_FORMATION_AND_MATURATION_OF_MRNA_TRANSCRIPT |
| Enrichment Score (ES) | -0.7034809 |
| Normalized Enrichment Score (NES) | -1.7657976 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0015020526 |
| FWER p-Value | 0.015 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TAF12 | 2537 16058 2338 | 770 | 3.303 | -0.0174 | No | ||
| 2 | CD2BP2 | 17621 | 1337 | 2.070 | -0.0328 | No | ||
| 3 | SFRS7 | 22889 | 1763 | 1.472 | -0.0449 | No | ||
| 4 | ELL | 18586 | 1828 | 1.381 | -0.0382 | No | ||
| 5 | TCEB2 | 12563 23102 7476 | 1843 | 1.361 | -0.0290 | No | ||
| 6 | PCBP2 | 9533 | 1883 | 1.314 | -0.0215 | No | ||
| 7 | SNRPA1 | 1373 18216 | 2234 | 0.991 | -0.0331 | No | ||
| 8 | TCEB3 | 15707 | 2243 | 0.987 | -0.0263 | No | ||
| 9 | THOC4 | 5710 10114 | 2571 | 0.724 | -0.0387 | No | ||
| 10 | SUPT4H1 | 9938 | 2592 | 0.705 | -0.0346 | No | ||
| 11 | CSTF2 | 24257 | 2997 | 0.456 | -0.0531 | No | ||
| 12 | SFRS3 | 5428 23312 | 3086 | 0.418 | -0.0547 | No | ||
| 13 | SF4 | 18593 | 3372 | 0.297 | -0.0680 | No | ||
| 14 | GTF2A1 | 2064 8187 8188 13512 | 3399 | 0.290 | -0.0672 | No | ||
| 15 | GTF2B | 10489 | 3469 | 0.268 | -0.0690 | No | ||
| 16 | TAF13 | 8288 | 3504 | 0.259 | -0.0689 | No | ||
| 17 | RNGTT | 10769 2354 6272 | 3641 | 0.224 | -0.0746 | No | ||
| 18 | CCNT1 | 22140 11607 | 3966 | 0.159 | -0.0910 | No | ||
| 19 | CSTF3 | 10449 6002 | 4295 | 0.111 | -0.1079 | No | ||
| 20 | SF3B1 | 13954 | 4344 | 0.106 | -0.1097 | No | ||
| 21 | POLR2I | 12839 | 4567 | 0.087 | -0.1210 | No | ||
| 22 | RBMX | 5367 9708 | 5144 | 0.058 | -0.1517 | No | ||
| 23 | U2AF2 | 5813 5812 5814 | 6448 | 0.026 | -0.2219 | No | ||
| 24 | RNPS1 | 9730 23361 | 7695 | 0.007 | -0.2892 | No | ||
| 25 | TAF9 | 3213 8433 | 8410 | -0.003 | -0.3277 | No | ||
| 26 | RBM5 | 13507 3016 | 8413 | -0.003 | -0.3278 | No | ||
| 27 | PCF11 | 121 | 8489 | -0.004 | -0.3318 | No | ||
| 28 | CCNT2 | 3993 13079 | 8564 | -0.005 | -0.3358 | No | ||
| 29 | PAPOLA | 5261 9585 | 8954 | -0.010 | -0.3567 | No | ||
| 30 | POLR2B | 16817 | 9561 | -0.017 | -0.3893 | No | ||
| 31 | SNRPE | 9843 | 10126 | -0.025 | -0.4196 | No | ||
| 32 | PABPN1 | 12018 | 10393 | -0.029 | -0.4337 | No | ||
| 33 | SNRPB2 | 5470 | 10695 | -0.034 | -0.4498 | No | ||
| 34 | TAF4 | 14319 | 10887 | -0.037 | -0.4598 | No | ||
| 35 | POLR2C | 9750 | 11135 | -0.042 | -0.4728 | No | ||
| 36 | CDK7 | 21365 | 11619 | -0.052 | -0.4985 | No | ||
| 37 | GTF2A2 | 10654 | 11993 | -0.061 | -0.5182 | No | ||
| 38 | ERCC2 | 1549 18363 3812 | 12015 | -0.061 | -0.5189 | No | ||
| 39 | GTF2H1 | 4069 18236 | 12248 | -0.068 | -0.5310 | No | ||
| 40 | SMC1A | 2572 24225 6279 10780 | 13683 | -0.136 | -0.6074 | No | ||
| 41 | MNAT1 | 9396 2161 | 14706 | -0.256 | -0.6607 | No | ||
| 42 | CCAR1 | 7454 | 14792 | -0.271 | -0.6633 | No | ||
| 43 | DNAJC8 | 12712 | 14961 | -0.308 | -0.6702 | No | ||
| 44 | SFRS1 | 8492 | 15244 | -0.391 | -0.6825 | No | ||
| 45 | TAF5 | 23833 5934 3759 | 15310 | -0.414 | -0.6830 | No | ||
| 46 | SNRPB | 9842 5469 2736 | 15571 | -0.515 | -0.6933 | No | ||
| 47 | TCEB1 | 13995 | 15726 | -0.591 | -0.6972 | No | ||
| 48 | SRRM1 | 6958 | 15782 | -0.618 | -0.6957 | No | ||
| 49 | SF3B2 | 23974 | 15888 | -0.682 | -0.6964 | No | ||
| 50 | POLR2A | 5394 | 16021 | -0.780 | -0.6978 | Yes | ||
| 51 | SFRS2 | 9807 20136 | 16049 | -0.798 | -0.6934 | Yes | ||
| 52 | NFX1 | 2475 | 16077 | -0.824 | -0.6888 | Yes | ||
| 53 | CSTF1 | 12515 | 16233 | -0.954 | -0.6902 | Yes | ||
| 54 | PRPF8 | 20780 1371 | 16257 | -0.972 | -0.6843 | Yes | ||
| 55 | FUS | 6135 | 16356 | -1.019 | -0.6821 | Yes | ||
| 56 | DHX9 | 8845 4608 3948 | 16367 | -1.024 | -0.6751 | Yes | ||
| 57 | SFRS9 | 16731 | 16466 | -1.109 | -0.6723 | Yes | ||
| 58 | SF3B4 | 22269 | 16492 | -1.131 | -0.6653 | Yes | ||
| 59 | SNRPA | 7007 11992 7008 | 16563 | -1.214 | -0.6602 | Yes | ||
| 60 | CDK9 | 14619 | 16575 | -1.224 | -0.6518 | Yes | ||
| 61 | TXNL4A | 6567 11329 | 16582 | -1.228 | -0.6432 | Yes | ||
| 62 | MAGOH | 2386 16148 | 16585 | -1.230 | -0.6343 | Yes | ||
| 63 | NUDT21 | 12665 | 16589 | -1.233 | -0.6254 | Yes | ||
| 64 | RNMT | 7501 | 16629 | -1.276 | -0.6181 | Yes | ||
| 65 | PTBP1 | 5303 | 16652 | -1.298 | -0.6098 | Yes | ||
| 66 | SNRPG | 12622 | 16664 | -1.317 | -0.6007 | Yes | ||
| 67 | SFRS5 | 9808 2062 | 16665 | -1.319 | -0.5911 | Yes | ||
| 68 | GTF2E1 | 22597 | 16701 | -1.364 | -0.5829 | Yes | ||
| 69 | RDBP | 6589 | 16730 | -1.400 | -0.5742 | Yes | ||
| 70 | TAF6 | 16322 891 | 16824 | -1.518 | -0.5681 | Yes | ||
| 71 | ERCC3 | 23605 | 16832 | -1.531 | -0.5572 | Yes | ||
| 72 | TAF11 | 12733 | 16854 | -1.553 | -0.5470 | Yes | ||
| 73 | GTF2E2 | 18635 | 16888 | -1.589 | -0.5371 | Yes | ||
| 74 | CCNH | 7322 | 16942 | -1.644 | -0.5279 | Yes | ||
| 75 | SF3A3 | 16091 | 17000 | -1.699 | -0.5185 | Yes | ||
| 76 | METTL3 | 21841 | 17006 | -1.704 | -0.5063 | Yes | ||
| 77 | TBP | 671 1554 | 17049 | -1.751 | -0.4957 | Yes | ||
| 78 | POLR2E | 3325 19699 | 17108 | -1.814 | -0.4856 | Yes | ||
| 79 | POLR2G | 23753 | 17143 | -1.872 | -0.4737 | Yes | ||
| 80 | PCBP1 | 17085 | 17187 | -1.942 | -0.4618 | Yes | ||
| 81 | TAF10 | 10785 | 17191 | -1.953 | -0.4476 | Yes | ||
| 82 | YBX1 | 5929 2389 | 17308 | -2.118 | -0.4383 | Yes | ||
| 83 | SF3A2 | 19938 | 17326 | -2.138 | -0.4236 | Yes | ||
| 84 | SF3A1 | 7450 | 17384 | -2.247 | -0.4102 | Yes | ||
| 85 | SNRPF | 7645 | 17393 | -2.259 | -0.3940 | Yes | ||
| 86 | GTF2F1 | 13599 | 17601 | -2.652 | -0.3858 | Yes | ||
| 87 | SFRS6 | 14751 | 17617 | -2.689 | -0.3669 | Yes | ||
| 88 | POLR2J | 16672 | 17714 | -2.890 | -0.3508 | Yes | ||
| 89 | SNRPD3 | 12514 | 17723 | -2.911 | -0.3299 | Yes | ||
| 90 | PHF5A | 22194 | 17755 | -2.970 | -0.3098 | Yes | ||
| 91 | CPSF1 | 22241 | 17788 | -3.055 | -0.2891 | Yes | ||
| 92 | SNRPD2 | 8412 | 17892 | -3.333 | -0.2703 | Yes | ||
| 93 | TH1L | 14715 | 17900 | -3.349 | -0.2461 | Yes | ||
| 94 | SF3B3 | 18746 | 17955 | -3.510 | -0.2233 | Yes | ||
| 95 | GTF2H3 | 5551 | 18028 | -3.729 | -0.1998 | Yes | ||
| 96 | U2AF1 | 8431 | 18097 | -3.895 | -0.1749 | Yes | ||
| 97 | PRPF4 | 7655 12845 | 18177 | -4.216 | -0.1483 | Yes | ||
| 98 | EFTUD2 | 1219 20203 | 18188 | -4.255 | -0.1176 | Yes | ||
| 99 | GTF2H2 | 6236 | 18200 | -4.311 | -0.0866 | Yes | ||
| 100 | NCBP2 | 12643 | 18234 | -4.445 | -0.0558 | Yes | ||
| 101 | POLR2H | 10888 | 18316 | -4.847 | -0.0246 | Yes | ||
| 102 | GTF2F2 | 21750 | 18429 | -5.557 | 0.0101 | Yes |