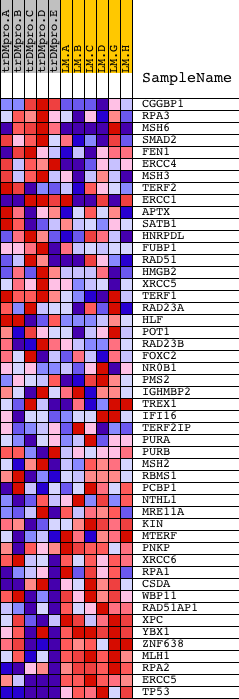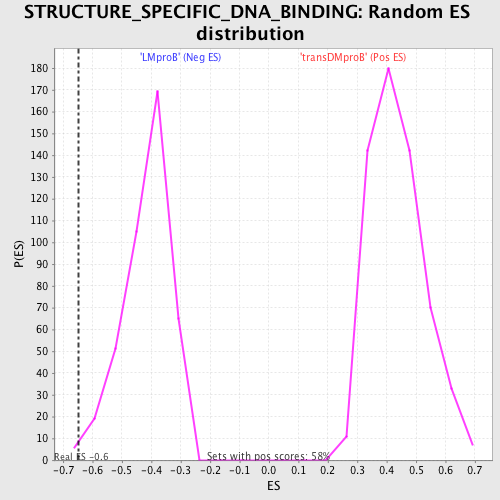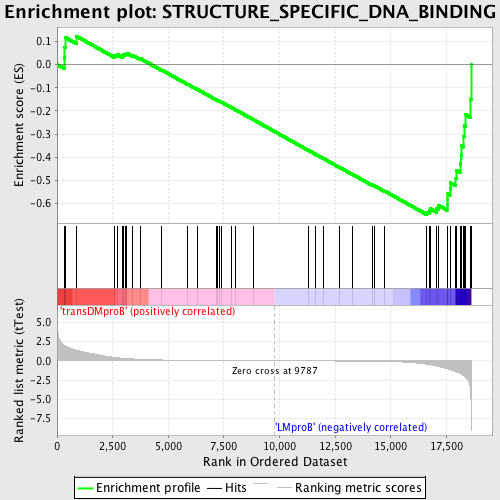
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | STRUCTURE_SPECIFIC_DNA_BINDING |
| Enrichment Score (ES) | -0.64767283 |
| Normalized Enrichment Score (NES) | -1.5514113 |
| Nominal p-value | 0.009638554 |
| FDR q-value | 0.61934954 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CGGBP1 | 22724 | 323 | 2.000 | 0.0296 | No | ||
| 2 | RPA3 | 12667 | 346 | 1.954 | 0.0744 | No | ||
| 3 | MSH6 | 23137 5120 | 380 | 1.889 | 0.1170 | No | ||
| 4 | SMAD2 | 23511 | 864 | 1.376 | 0.1234 | No | ||
| 5 | FEN1 | 8961 | 2585 | 0.433 | 0.0409 | No | ||
| 6 | ERCC4 | 6927 11921 | 2702 | 0.393 | 0.0439 | No | ||
| 7 | MSH3 | 5119 | 2956 | 0.321 | 0.0378 | No | ||
| 8 | TERF2 | 18756 | 2969 | 0.317 | 0.0446 | No | ||
| 9 | ERCC1 | 1235 1045 1646 | 3093 | 0.287 | 0.0448 | No | ||
| 10 | APTX | 2455 7291 | 3114 | 0.283 | 0.0503 | No | ||
| 11 | SATB1 | 5408 | 3397 | 0.230 | 0.0406 | No | ||
| 12 | HNRPDL | 11947 3551 6954 | 3730 | 0.185 | 0.0270 | No | ||
| 13 | FUBP1 | 15390 | 4711 | 0.100 | -0.0234 | No | ||
| 14 | RAD51 | 2897 14903 | 5880 | 0.057 | -0.0850 | No | ||
| 15 | HMGB2 | 13594 | 6290 | 0.047 | -0.1059 | No | ||
| 16 | XRCC5 | 14229 | 7184 | 0.032 | -0.1532 | No | ||
| 17 | TERF1 | 14293 | 7215 | 0.031 | -0.1541 | No | ||
| 18 | RAD23A | 3878 5348 18813 9685 | 7300 | 0.030 | -0.1579 | No | ||
| 19 | HLF | 10124 | 7395 | 0.028 | -0.1623 | No | ||
| 20 | POT1 | 17201 | 7854 | 0.022 | -0.1865 | No | ||
| 21 | RAD23B | 5349 | 8038 | 0.019 | -0.1959 | No | ||
| 22 | FOXC2 | 18442 | 8818 | 0.011 | -0.2376 | No | ||
| 23 | NR0B1 | 24291 | 11315 | -0.016 | -0.3717 | No | ||
| 24 | PMS2 | 16632 | 11592 | -0.020 | -0.3860 | No | ||
| 25 | IGHMBP2 | 23945 | 11995 | -0.025 | -0.4071 | No | ||
| 26 | TREX1 | 10219 3111 | 12675 | -0.035 | -0.4429 | No | ||
| 27 | IFI16 | 13745 | 13274 | -0.046 | -0.4740 | No | ||
| 28 | TERF2IP | 12184 18463 7167 | 14154 | -0.069 | -0.5197 | No | ||
| 29 | PURA | 9670 | 14274 | -0.073 | -0.5244 | No | ||
| 30 | PURB | 5335 | 14734 | -0.092 | -0.5470 | No | ||
| 31 | MSH2 | 23138 | 16605 | -0.441 | -0.6373 | Yes | ||
| 32 | RBMS1 | 14580 | 16721 | -0.499 | -0.6318 | Yes | ||
| 33 | PCBP1 | 17085 | 16779 | -0.526 | -0.6225 | Yes | ||
| 34 | NTHL1 | 23357 | 17063 | -0.685 | -0.6216 | Yes | ||
| 35 | MRE11A | 3014 19560 | 17156 | -0.746 | -0.6090 | Yes | ||
| 36 | KIN | 15120 | 17528 | -1.017 | -0.6051 | Yes | ||
| 37 | MTERF | 9911 | 17533 | -1.020 | -0.5813 | Yes | ||
| 38 | PNKP | 4052 18258 | 17540 | -1.028 | -0.5575 | Yes | ||
| 39 | XRCC6 | 2286 8991 | 17660 | -1.136 | -0.5372 | Yes | ||
| 40 | RPA1 | 20349 | 17680 | -1.170 | -0.5107 | Yes | ||
| 41 | CSDA | 1163 16966 1134 | 17927 | -1.415 | -0.4906 | Yes | ||
| 42 | WBP11 | 16953 | 17945 | -1.435 | -0.4578 | Yes | ||
| 43 | RAD51AP1 | 5350 1073 | 18117 | -1.643 | -0.4284 | Yes | ||
| 44 | XPC | 5925 | 18154 | -1.700 | -0.3904 | Yes | ||
| 45 | YBX1 | 5929 2389 | 18178 | -1.746 | -0.3505 | Yes | ||
| 46 | ZNF638 | 9476 | 18282 | -1.954 | -0.3101 | Yes | ||
| 47 | MLH1 | 18982 | 18301 | -2.002 | -0.2640 | Yes | ||
| 48 | RPA2 | 2330 16057 | 18374 | -2.174 | -0.2168 | Yes | ||
| 49 | ERCC5 | 14255 4097 | 18568 | -3.343 | -0.1486 | Yes | ||
| 50 | TP53 | 20822 | 18610 | -6.426 | 0.0003 | Yes |